
μCaler HotSpot Panel v1.0
#1101401
μCaler HotSpot Panel v1.0 targets the“hotspot" regions of tumor mutation, involving 49 genes related to carcinogenesis and tumor suppressor, with probes covering approximately 25 Kb of the genome.
This Panel is designed for use with the μCaler Hybrid Capture System. Based on the novel μCaler targeted enrichment system, the μCaler HotSpot Panel v1.0 delivers improved capture efficiency, consistent and excellent capture performance, and sig- nificantly reduces experimental time, enabling more efficient sequencing and cost savings.
- Product details
- Kit Content
- FAQ
- Ordering Information
- Resources
Gene list
| AKT1 | ALK | APC | AR | ARAF | ATM | BRAF | CDKN2A | CHEK2 | CTNNB1 |
| DDR2 | EGFR | ERBB2 | ERBB3 | ESR1 | FBXW7 | FGFR1 | FGFR2 | FGFR3 | FGFR4 |
| FLT3 | GNA11 | GNAQ | GNAS | HRAS | IDH1 | IDH2 | KIT | KRAS | MAP2K1 |
| MAP2K2 | MET | MTOR | NRAS | NTRK1 | NTRK3 | PDGFRA | PIK3CA | PTCH1 | PTEN |
| RAF1 | RET | ROS1 | SF3B1 | SMAD4 | SMO | STK11 | TP53 | TSC1 |
Performance
Capture Performance
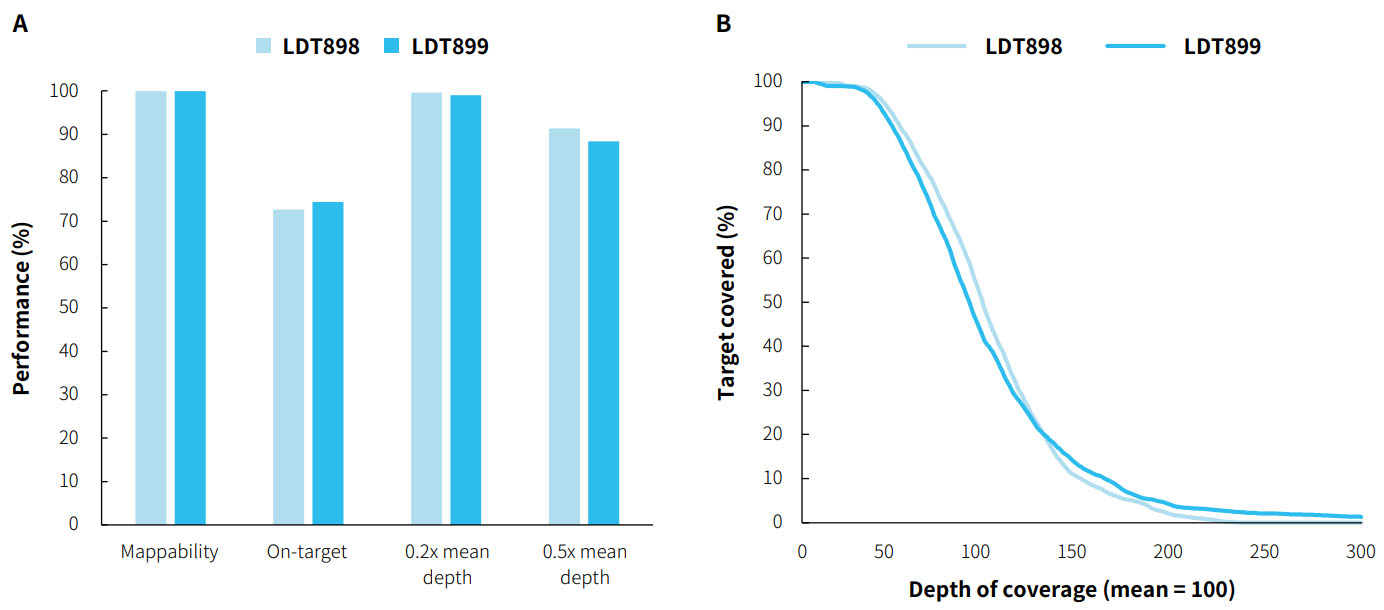
Fig 1. Capture performance of μCaler HotSpot Panel v1.0. LDT OncoOne Pan-tumor gDNA Standard (LDT Bioscience, LDT900) was used for pre-library preparation with NadPrep® EZ DNA Library Preparation Kit v2 paired with NadPrep® Universal Stubby Adapter (UDI) Module; 500 ng/pre-library (1-plex) input was used, following the user manual of μCaler hybrid capture. Sequencing was performed using Illumina Novaseq 6000, PE150. The BWA was used for alignment to the reference genome hg38 and On-target rate was calculated by the number of reads. A. Mappability, On-target rate and Target covered;B. Coverage uniformity.
Note:OncoOne Pan-cancer gDNA Standard (LDT Bioscience, LDT900) include a negative control (LDT Bioscience, LDT898) and a positive control (LDT Bioscience, LDT899).
Variant analysis
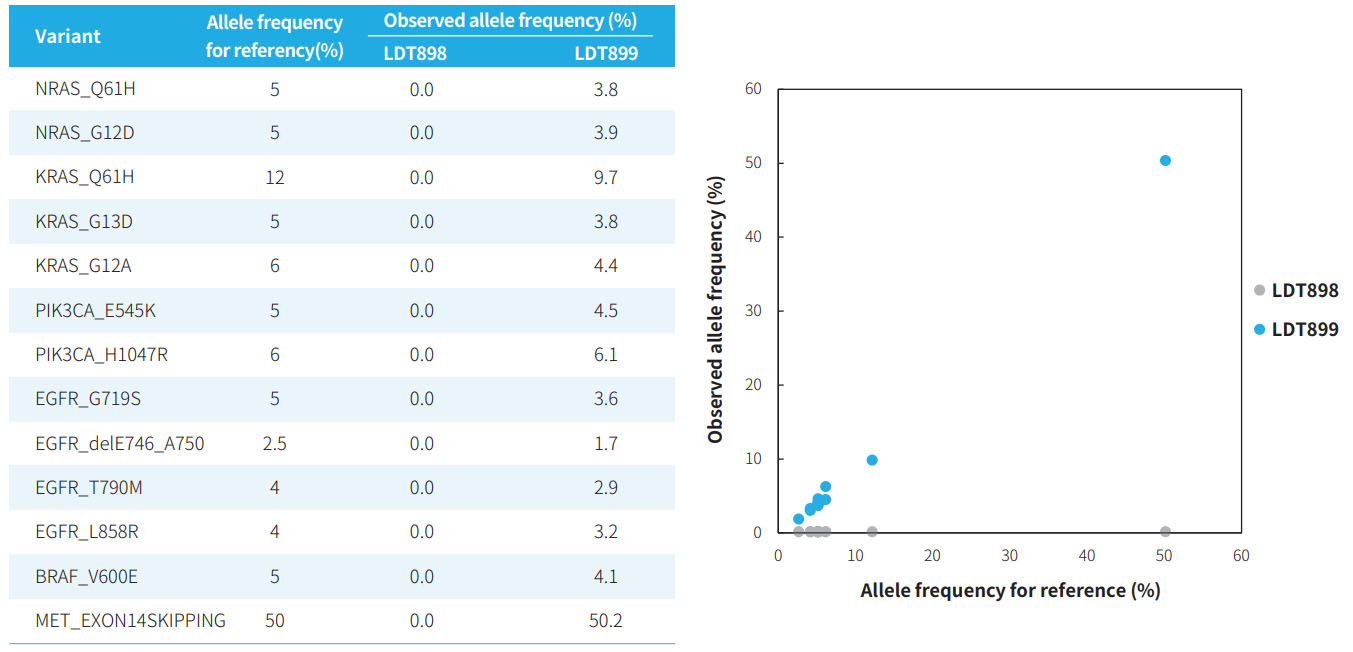
Fig 2. Consistency of the allele frequency in μCaler HotSpot Panel v1.0 capture data with the reference frequency of the standards.Pre-libraries were prepared using the NadPrep EZ DNA Library Preparation Kit v2, μCaler HotSpot Panel v1.0 was used to complete hybridization capture. Sequencing was performed using Illumina Novaseq 6000, PE150.
Note:The samples were: OncoOne Pan-cancer gDNA Standard (LDT Bioscience, LDT900).
For research use only. Not for use in diagnostic procedures.
|
Catalog |
Color of Tube Cap |
Item |
Volume |
Package/Storage |
|
1101402 |
|
μCaler HotSpot Panel v1.0, 16 rxn |
40 μL |
-20℃ |
|
1101401 |
|
μCaler HotSpot Panel v1.0, 96 rxn |
210 μL |
-20℃ |
|
Product |
Catalog |
|
μCaler HotSpot Panel v1.0, 16 rxn |
1101402 |
|
μCaler HotSpot Panel v1.0, 96 rxn |
1101401 |
Product Sheet
-
[Product Catalog] -
Product Catalog
Download
-
[Technical Note] -
EN_Protocol_μCaler Hybrid Capture of DNA Library (for Illumina®) v2.0
Download
-
[Technical Note] -
EN_Protocol_μCaler Hybridization Capture of DNA Library (for MGI) v1.0
Download
-
[Technical Note] -
EN_Checklist_μCaler Hybrid Capture of DNA Libraries (for Illumina)
Download
-
[Poster] -
EN_Brochure_μCaler HotSpot Panel v1.0
Download
Application Note
Demo Data
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)

Nanodigmbio will contact you within 1 day.
If there are any problems, please contact us by 400 8717 699 / support@njnad.com

