Methyl Library Preparation Total Solution
Background
DNA methylation, an epigenetic modification, is closely related to the gene expression and chromatin remodeling. The precise identification of cytosine methylation status in genome plays a critical role in research and clinical areas, especially in precise oncology. With the treatment of bisulfite or enzymes, the unmethylated cytosine can be converted to uracil, whereas the methylated cytosine remains unchanged. Afterwards, combined with high-throughput sequencing technology, the genomic methylation status can be analyzed massively with single nucleotide resolution. The introduction of targeted enrichment technology can reduce the sequencing cost and improve the targeted analysis efficiency by improving the sequencing depth of interested regions. Nanodigmbio offers an NGS total solution for capture-based methylation sequencing (Methyl-Seq) on both MGI sequencing platforms and Illumina sequencing platforms, including Methyl-Seq library preparation kit based on bisulfite or enzymatic conversion, design & synthesis service of capture probes, hybridization reagents related and integrated analysis tools.
Solution
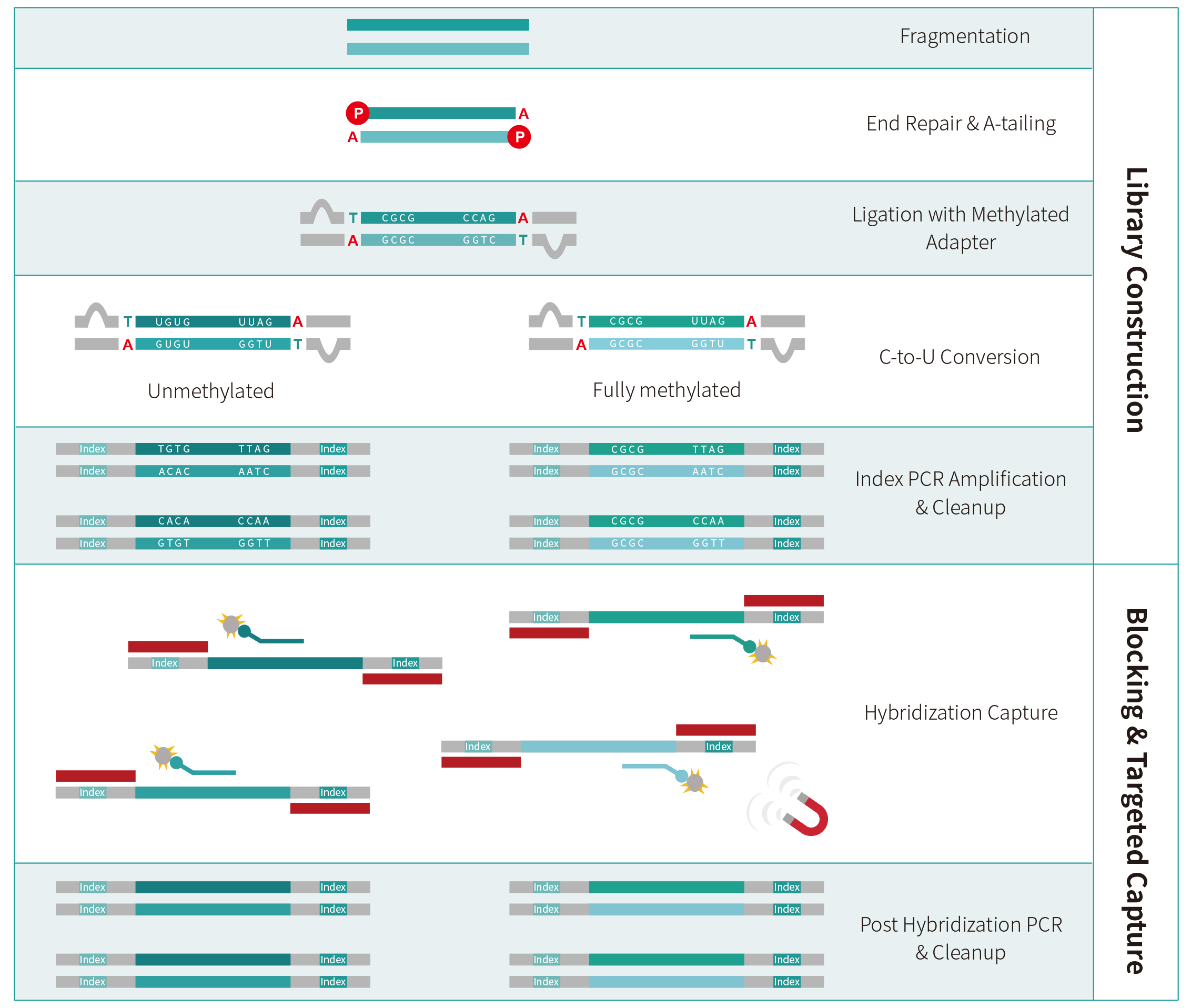
|
Library Preparation |
Blocking |
Target Capture |
|
panels NadPrep NadPrep Hybrid Capture Reagents |
||
|
NadPrep Methyl Library Preparation Module (for Illumina®️)
NadPrep Methyl UDI Adapter Kit (for Illumina®️)
|
panels NadPrep Hybrid Capture Reagents |
Performance
Library Yield and Conversion Efficiency
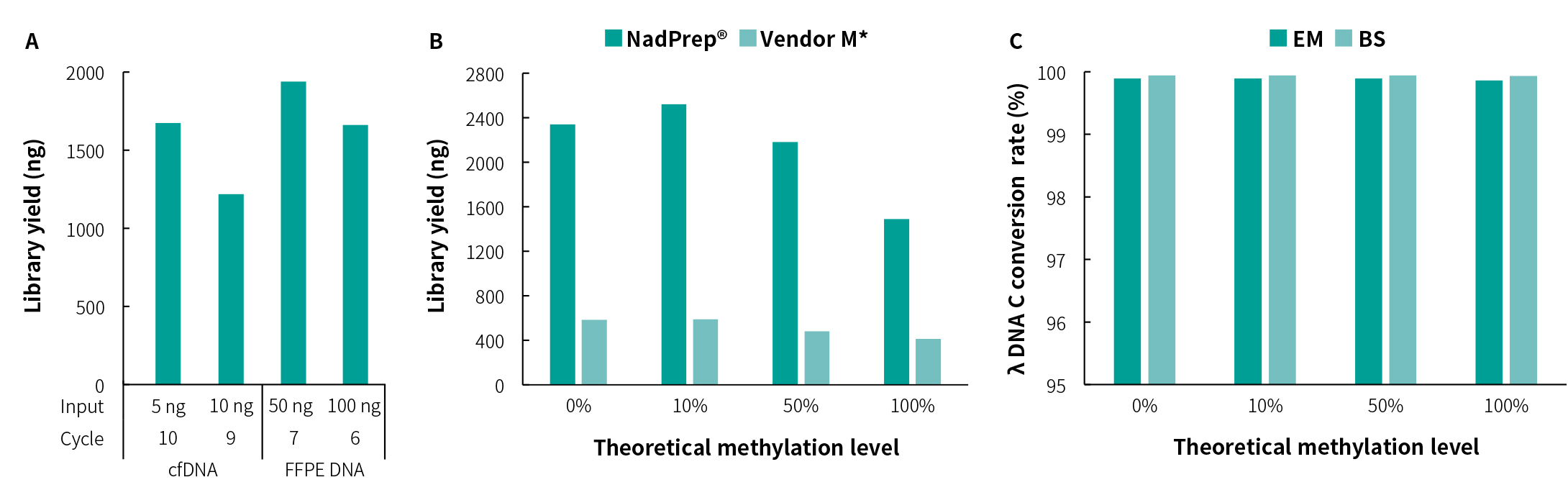
Fig 1.Library preparation performance of different types of samples and different conversion methods. A. Library yield of different types of samples,B. Library yield and C. λ DNA C conversion rate of of simulated samples with different methylation levels indicate stable performance and differences in conversion step treated by enzymatic (EM) and bisulfite (BS), respectively. The libraries were prepared by using NadPrep Methyl Library Preparation Module coupled with NadPrep Methyl Adapter (SI/MDI) Module (for MGI).
Note:The human 100% methylated DNA standard (Zymo, D5014-2) positive control and 0% methylated DNA standard (Zymo, D5014-1) negative control were mixed in different proportions to simulate different methylation levels (0%, 10%, 50% and 100%).
Capture Performance

Fig 2. Capture performance of libraries treated by different conversion methods.A. Mappability and On-target rate, B. Target covered, C. Coverage uniformity and consistency, and D. GC bias show satisfying performance treated by EM and BS, respectively.Note: Samples are simulated DNA by using human 100% methylated DNA standard (zymo, D5014-2) positive control and 0% methylated DNA standard (zymo, D5014-1) negative control to mimic different methylation levels (0%, 10%, 50% and 100%).
Methylation Detection
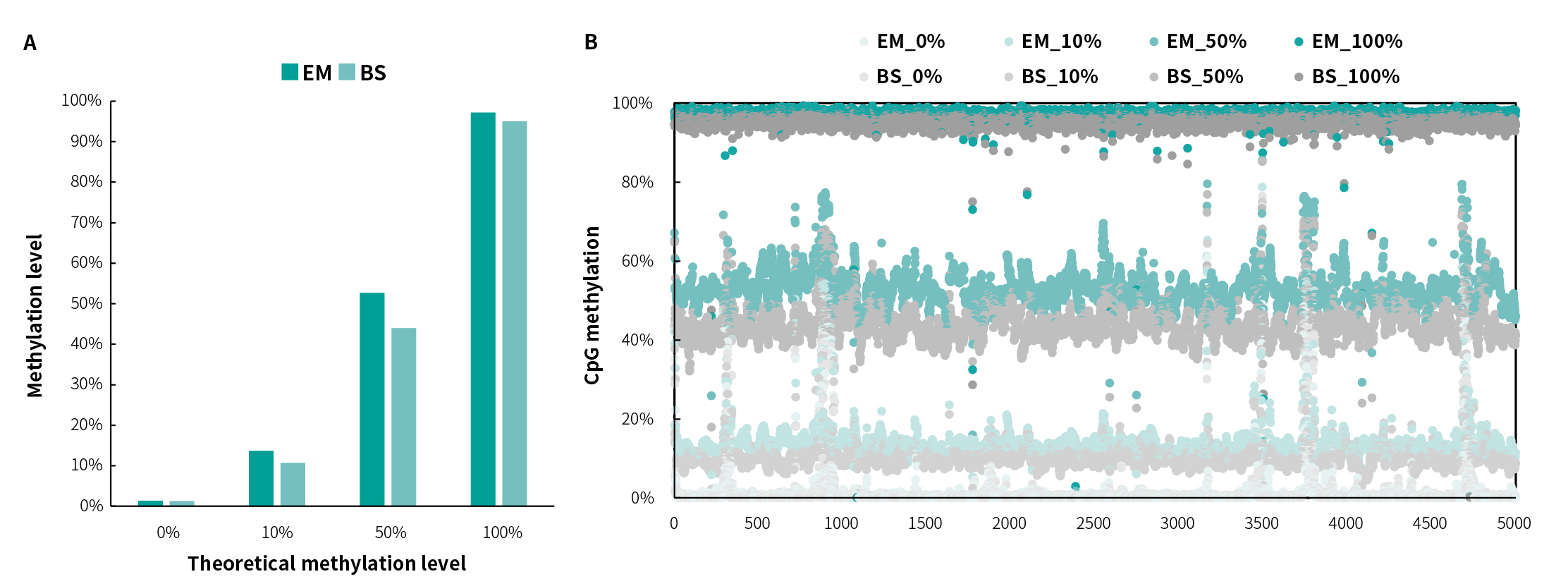
Fig 3. NadPrep Performance of different methylation level samples treated by EM and BS solutions. The libraries were prepared by using NadPrep Methylation Library Preparation Module coupled with NadPrep Methyl Adapter (MDI) Module (for MGI). The capture was applied by a methylation demo panel (60 Kb), then sequenced on MGISEQ-2000, PE100. A. The actual detection results of samples with different methylation levels and B. The CpG methylation level of samples treated by EM and BS conversion solution are shown, respectively.Note: Samples are simulated DNA by using human 100% methylated DNA standard (zymo, D5014-2) positive control and 0% methylated DNA standard (zymo, D5014-1) negative control to mimic different methylation levels (0%, 10%, 50% and 100%).
Library Yield and Conversion Efficiency

Fig 4.Library preparation performance of different conversion methods. The libraries were prepared by using NadPrep Methylation Library Preparation
Module coupled with NadPrep Methyl Stubby Adapter (UDI) Module (for Illumina®) (Stubby) and NadPrep Methyl UDI Adapter Module (for Illumina®) (Full)
in conversion step treated by enzymatic (EM) and bisulfite (BS), respectively. A. Library yield of two kinds of adapters in different conversion step. B. Library A
yield of two kinds of stubby adapters in conversion step treated by EM. C. λ DNA C conversion rate in different conversion step.
Note:Samples are simulated DNA samples by using human 100% methylated DNA standard (zymo, D5014-2) positive control and 0% methylated DNA
T
standard (zymo, D5014-1) negative control to mimic different methylation levels (0%, 50% and 100%). Input amount: 50 ng. The adapter of B & C are Stubby.
Capture Performance

Fig5.Capture performance of libraries treated by different conversion methods.The libraries were prepared by using NadPrep Methylation Library Preparation Module coupled with NadPrep Methyl Stubby Adapter (UDI) Module (for Illumina®). The capture was applied by a methylation demo panel (60 Kb), then sequenced on Novaseq 6000, PE 150. The A. Mappability and On-target rate, B. Target covered, C. Coverage uniformity and consistency, and D. GC bias show satisfying performance treated by EM and BS, respectively.
Methylation Detection
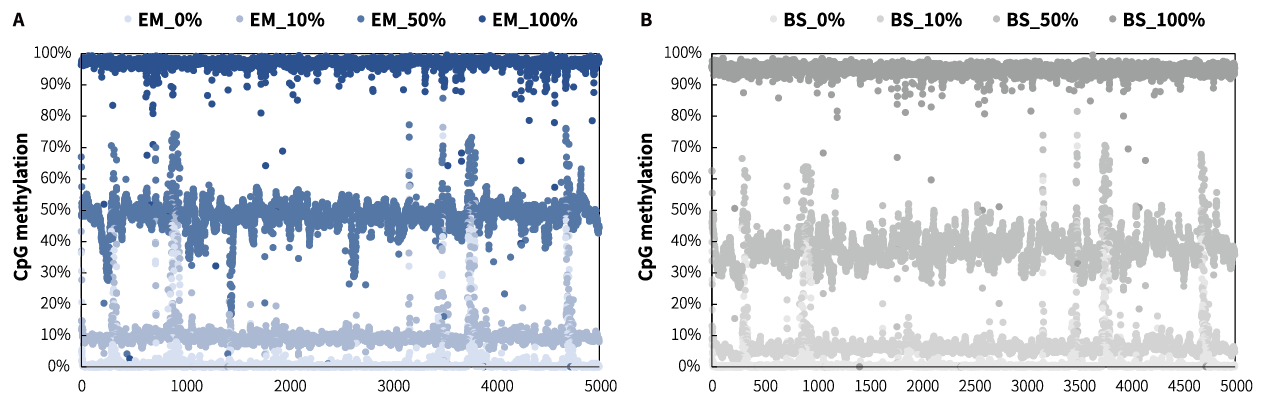
Fig 6. Performance of different methylation level samples treated by EM and BS solutions. The libraries were prepared by using NadPrep Methylation Library Preparation Module coupled with NadPrep Methyl Stubby Adapter (UDI) Module (for Illumina®). The capture was applied by a methylation demo panel (60 Kb), then sequenced on Novaseq 6000, PE 150. A. The CpG methylation level of samples treated by EM conversion solution; B. The CpG methylation level of samples treated by BS conversion solution.
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)


