RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
Background
NGS technology plays an important role during the battle against SARS-CoV-2. In recent years, with rapid development and decreased cost, NGS technology is becoming routinely employed in respiratory virus detection.
One of the challenges for NGS as a virus detection and discovery tool is the low abundance of viral nucleic acids in relation to host in clinical samples. Several methods have been applied to remove host cells and exogenous nucleic acid to increase the proportion of virus sequences obtained by NGS, including ultracentrifugation, filtration, nuclease treatments and rRNA depletion. Although these methods are helpful, they may not achieve the desired sensitivity[1]. To address this problem, targeted NGS has been implemented to increase the sensitivity of sequence-based virus detection and characterization.
NanoRV Panel v1.0-Tech Access (referred to as NanoRV Panel in the following) developed by Nanodigmbio is a hybridization capture panel targeting multiple respiratory viruses including Coronavirus (CoVs) (Fig 1).
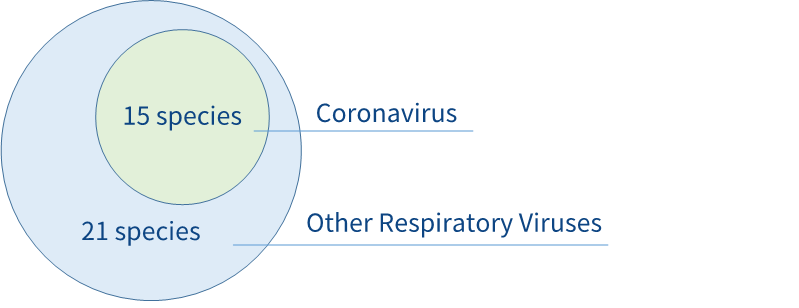
Fig 1. Targets of the NanoRV Panel.The panel was designed to target human respiratory viruses, including coronavirus (with SARS-CoV-2 considered) and other respiratory viruses, e.g., influenza/parainfluenza virus, adenovirus, orthopneumovirus, metapneumovirus, enterovirus, rhinovirus and bocaparvovirus.
Compared to amplicon sequencing, hybridization capture exhibits much better sequence variation tolerance and is readily expandable. This is especially important for RNA virus detection. For the lack of proofreading ability in RNA polymerases, RNA viruses are prone to mutation introduced during replicating process. CoVs have almost the largest nonsegmented genomes (30 kb) among all RNA viruses. Large genomes enhance plasticity, which in turn leads to greater diversity[2]. The real-time surveillance suggested that SARS-CoV-2 has accumulated mutations at a rate of 1 - 2 changes / month. Some occurred in the gene that encodes the spike protein on the viral surface[3]. Multiple SARS-CoV-2 variants are circulating globally, including most notably the B.1.1.7 lineage that emerged in the United Kingdom, and B.1.135 in South Africa[4].
With these new variants emerging, an amplicon-based viral genome enrichment scheme would need to be regularly updated. However, a hybridization capture based panel should remain efficient. For the 120 bp target region covered by one probe, it will take on average decades to accumulate enough mutations beyond the probe’s tolerance. Even though mutations accumulating, the target sequences can also be acquired as flanking regions captured by neighboring probes (Fig 2).
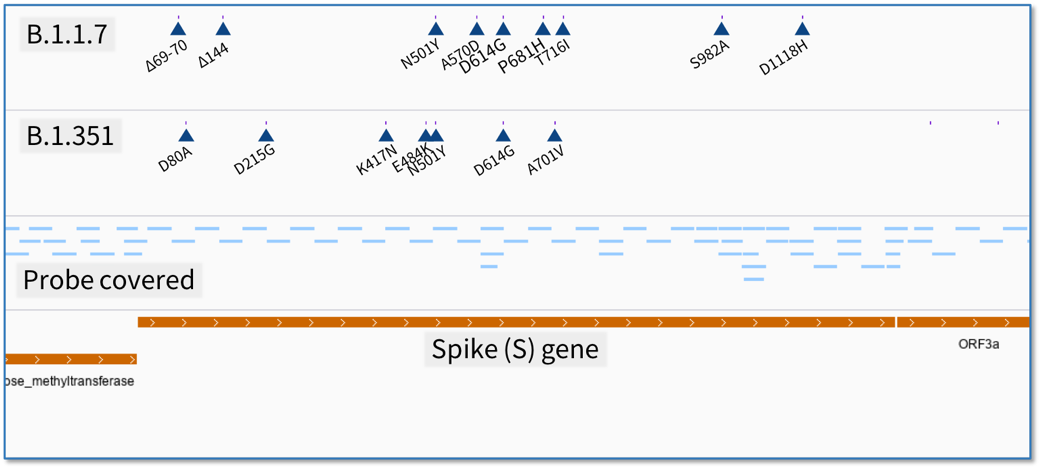
Fig 2. Mutations distribution and probe covering over the spike gene of B.1.1.7 and B.1.351.
Nanodigmbio provides a reliable solution for RNA-Cap Sequencing of RNA viruses such as SARS-CoV-2. One can choose either single capture or double capture workflows according to priority and sample characteristic.
Solution

Fig 3. Workflow and reagents.
|
cDNA Synthesis |
Library Preparation |
Blocking |
Target Capture |
|
NadPrep Total RNA-To-DNA Module
|
NadPrep DNA Universal Library Preparation Kit (for MGI)
|
NadPrep NanoBlockers (for MGI)
|
|
|
NadPrep DNA Universal Library Preparation Kit (for Illumina®)
|
NadPrep Hybrid Capture Reagents
|
Performance
The libraries are prepared from COVID-19 RNA reference (Genewell, GW-CRPM002) composed of complete SARS-CoV-2 genome and human RNA. 20 ng RNA samples (containing ~ 267 copies of SARS-CoV-2 genome) were reverse-transcribed into cDNA using the NadPrep Total RNA-To-DNA Module, then the DNA library construction were completed with NadPrep DNA Universal Library Preparation Kit (for Illumina®). Custom probes targeting 7 human housekeeping genes were spiked into the NanoRV Panel, followed by either single or double capture sequencing. Sequencing data was analyzed with Salmon v1.2.0. Reads mapping to SARS-CoV-2 or 7 housekeeping genes were counted as on-target, while to rRNAs or other transcripts as off-target.
(1) On-target Rate
Double capture increased the on-target rate from 56.3% to 99.5% with rRNA mostly depleted.
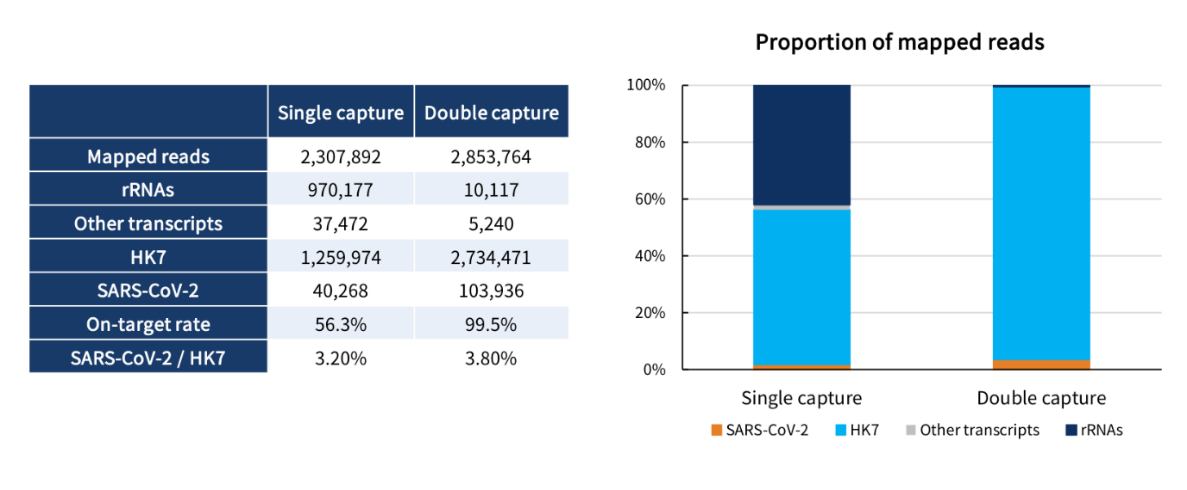
Fig 4. On-target rate of single capture and double capture.
(2) Relative RNA Abundance
Single capture and double capture exhibited similar trendlines of the RNA abundance for SARS-CoV-2 and 7 housekeeping genes.
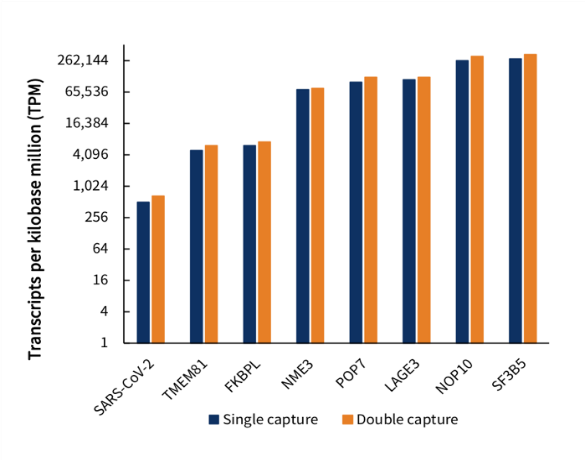
Fig 5. RNA abundance of SARS-CoV-2 and 7 housekeeping genes for single capture and double capture.
(3) Coverage Depth and Uniformity
> 99% target regions of the SARS-CoV-2 reached 0.2x mean depth of coverage. Double capture yielded more raw viral data with coverage uniformity slightly down.
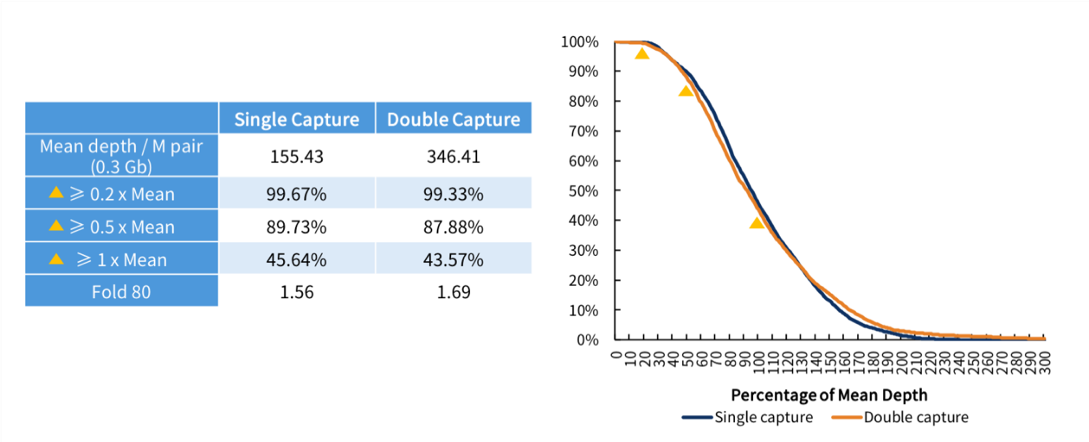
Fig 6. Coverage uniformity of SARS-CoV-2 for single capture and double capture.
Nanodigmbio provides a reliable solution for RNA-Cap Sequencing of RNA viruses such as SARS-CoV-2. One can choose either single capture or double capture workflows according to priority and sample characteristic.
References
[1] O'Flaherty, Brigid M., et al. Comprehensive viral enrichment enables sensitive respiratory virus genomic identification and analysis by next generation sequencing. Genome research 28.6 (2018): 869-877.
[2] Li, Bei, et al. Discovery of bat coronaviruses through surveillance and probe capture-based next-generation sequencing. Msphere 5.1 (2020).
[3] https://www.sciencemag.org/news
[4] https://www.cdc.gov/coronavirus/
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)


