
HiSNP-series Panel
#1001838 (HiSNP Ultra Panel v1.0, 960 rxn)
#1001839 (HiSNP Ultra Panel v1.0, 1920 rxn)
HiSNP Ultra Panel v1.0 is designed for analyzing genomic composition status with high resolution. This SNP skeleton-based panel can scan the whole genome with 50 Kb apart by targeting over 52,000 high MAF SNP sites.
HiSNP Panel v1.0 is designed for analyzing genomic composition status with basic resolution. This SNP skeleton-based panel can scan the whole genome with 300 Kb apart by targeting over 9,000 high MAF SNP sites.
- Product details
- Kit Content
- FAQ
- Ordering Information
- Resources
Feature
- High heterozygosity ratio: Comprehensive designing for Europeans, Africans, Americans and Asians (AF=0.05-0.95)
- Stable capture performance: GC balanced; unique flanking sequence (verified by WGS)
- Appropriate proportionality: Even distribution in whole genome; LD sites excluded
- Superior compatibility: Use alone or spike-in other CDS panels
|
Product |
Application |
|
HiSNP Ultra Panel v1.0 |
HRD Score analysis
CNV
ID |
|
HiSNP Panel v1.0 |
CNV
ID |
Performance
Capture performance of HiSNP-series Panel
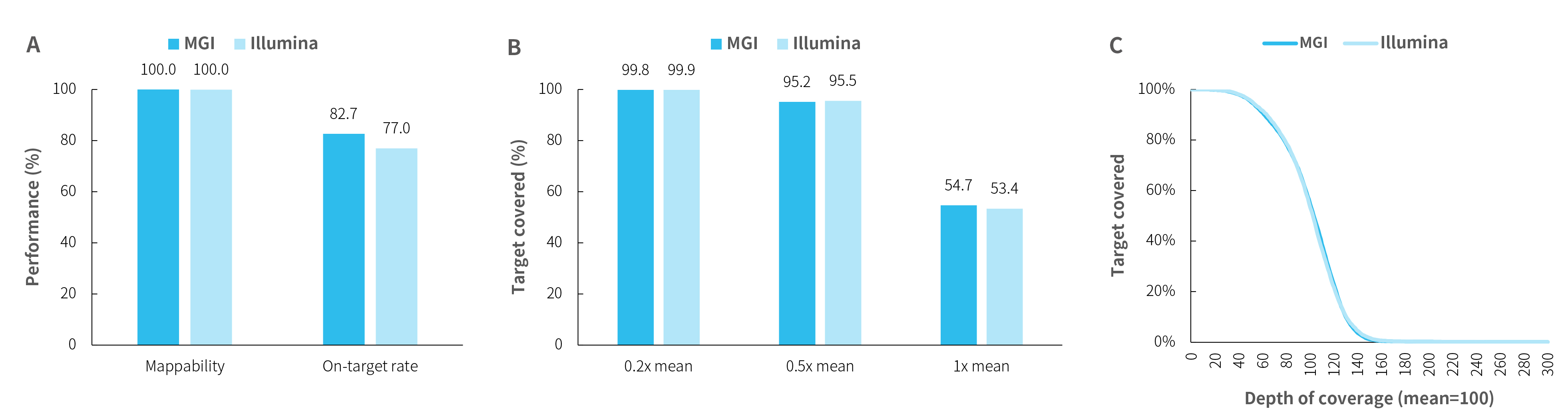
Fig 1. Capture performance of HiSNP-series Panel. DNA libraries were prepared using the NadPrep DNA Library Preparation Kit (for MGI) and the NadPrep DNA Library Preparation Kit (for Illumina®), respectively. A. mapping rate, on-target rate;B-C. coverage uniformity and consistency of HiSNP (HiSNP Panel v1.0) and HiSNP Ultra (HiSNP Ultra Panel v1.0) were exhibited, respectively.
Note:Sample type: human genomic DNA, male (Promega, G1471). MGI platform: MGISEQ-2000 with PE100; Illumina platform: NovaSeq 6000 with PE150.
Capture performance of HiSNP Ultra Panel v1.0
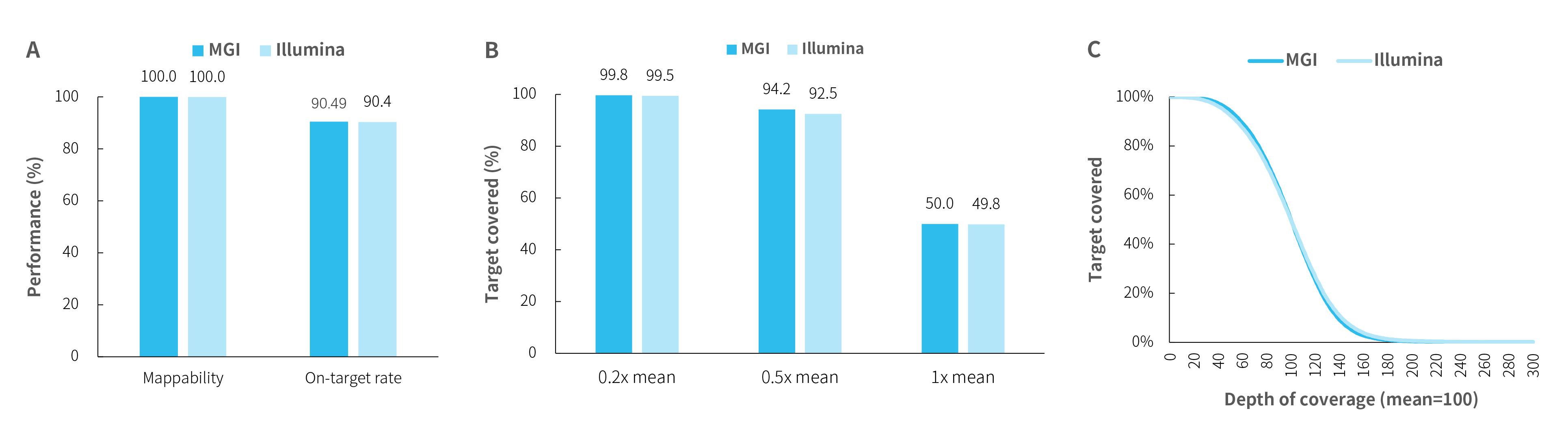
Fig 2. Capture performance of HiSNP Ultra Panel v1.0 with HRD standards. DNA libraries were prepared using the NadPrep DNA Library Preparation Kit (for MGI) and the NadPrep DNA Library Preparation Kit (for Illumina®), respectively. A. mapping rate, on-target rat;B-C. coverage uniformity and consistency of HiSNP (HiSNP Panel v1.0) and HiSNP Ultra (HiSNP Ultra Panel v1.0) were exhibited, respectively.。
Note:Sample type: human genomic DNA, male (Promega, G1471). MGI platform: MGISEQ-2000 with PE100; Illumina platform: NovaSeq 6000 with PE150.
Capture Performance of HiSNP Ultra Panel v1.0 with HRD Standard
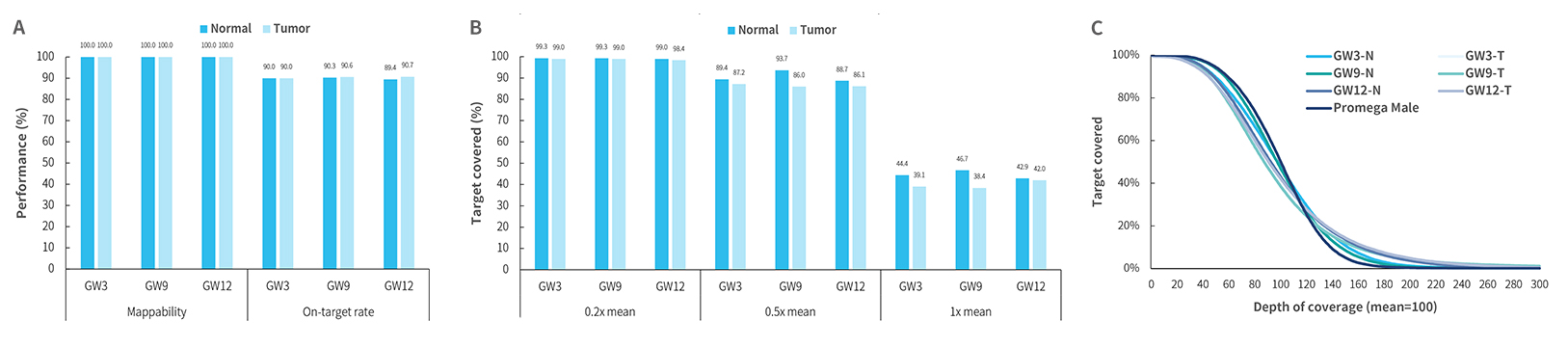
Fig3. Capture performance of HiSNP Ultra Panel v1.0 with HRD standards. DNA libraries were prepared using the NadPrep DNA Library Preparation Kit (for MGI) A. mapping rate, on-target rat;B-C. coverage uniformity and consistency of HiSNP (HiSNP Panel v1.0) and HiSNP Ultra (HiSNP Ultra Panel v1.0) were exhibited, respectively.
Note:GW3: GW-HRD-3 standards (Genewell); GW9: GW-HRD-9 standards (Genewell); GW12: GW-HRD-12 standards (Genewell). All standards are paired cell lines, including Normal (N) and Tumor (T). MGI platform: MGISEQ-2000 with PE100; Illumina platform: NovaSeq 6000 with PE150.
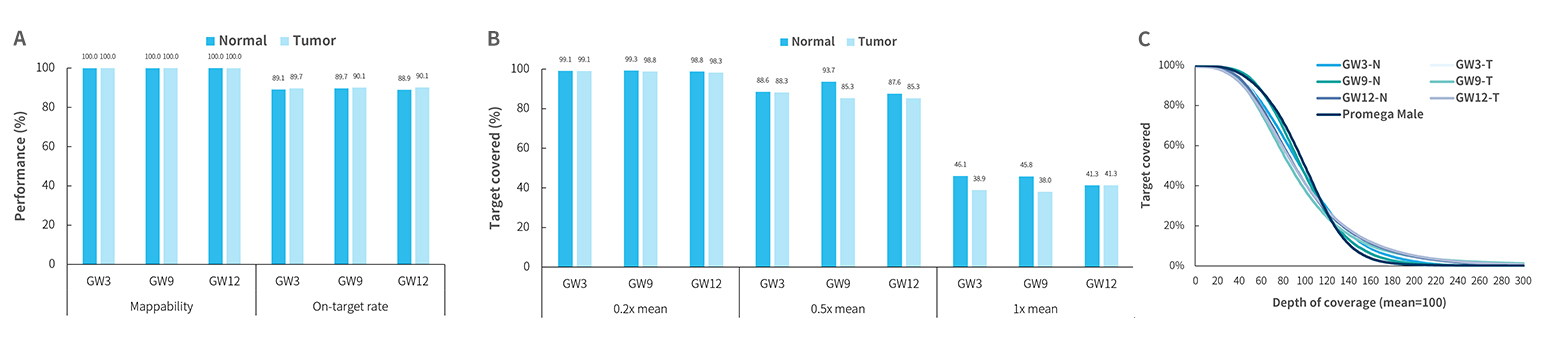
Fig4. Capture performance of HiSNP Ultra Panel v1.0 with HRD standards. DNA libraries were prepared using the NadPrep DNA Library Preparation Kit (for Illumina®) A. mapping rate, on-target rat;B-C. coverage uniformity and consistency of HiSNP (HiSNP Panel v1.0) and HiSNP Ultra (HiSNP Ultra Panel v1.0) were exhibited, respectively.
Note:Sample type: HRD standards (Genewell).GW3: GW-HRD-3 standards (Genewell); GW9: GW-HRD-9 standards (Genewell); GW12: GW-HRD-12 standards (Genewell). All standards are paired cell lines, including Normal (N) and Tumor (T). MGI platform: MGISEQ-2000 with PE100; Illumina platform: NovaSeq 6000 with PE150.
HiSNP for HRD Score

Fig 5. HRD score analysis of HiSNP Ultra Panel v1.0 with HRD standards by different platform. A. MGI Platform;B. Illumina Platform
Note: Sample type: HRD standards (Genewell).GW3: GW-HRD-3 standards (Genewell); GW9: GW-HRD-9 standards (Genewell); GW12: GW-HRD-12 standards (Genewell). All standards are paired cell lines, including Normal (N) and Tumor (T). MGI platform: MGISEQ-2000 with PE100; Illumina platform: NovaSeq 6000 with PE150.
For research use only. Not for use in diagnostic procedures.
| Catalog# | Color Of Tube Cap | Item | Volume | Package/Storage |
| 1001831 |
|
HiSNP Panel v1.0, 96 rxn | 415 μL | –20℃ |
| 1001838 |
|
HiSNP Ultra Panel v1.0, 960 rxn | 10×96 rxn(415 μL) | –20℃ |
| 1001839 |
|
HiSNP Ultra Panel v1.0, 1920 rxn | 20×96 rxn(415 μL) | –20℃ |
To spike-in use with other panels, please contact Nanodigmbio for technical support.
The sequencing depth of HiSNP Ultra is 75× per Gb theoretically. If you want to get 500× of sequencing depth, it is recommended to achieve 7.5 Gb / library.
| Product |
Details |
Catalog |
|
HiSNP Panel v1.0, 96 rxn |
96 rxn |
1001831 |
|
HiSNP Ultra Panel v1.0, 960 rxn |
960 rxn |
1001838 |
|
HiSNP Ultra Panel v1.0, 1920 rxn |
1920 rxn |
1001839 |
Product Sheet
-
[Product Catalog] -
Product Catalog
Download
-
[Technical Note] -
Hybridization Capture of DNA Libraries (for MGI) Protocol
Download
-
[Technical Note] -
Hybridization Capture of DNA Libraries (for for Illumina®) Protocol
Download
-
[Technical Note] -
HiSNP Ultra Panel v1.0_Instruction
Download
-
[Poster] -
HiSNP-series_Panels EN_Brochure__Version202106
Download
-
[Bed File] -
HiSNP Panel v1.0
Download
Application Note
Demo Data
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)

Nanodigmbio will contact you within 1 day.
If there are any problems, please contact us by 400 8717 699 / support@njnad.com


