
NEXome Plus Panel v1.0
#1001861
NEXome Plus Panel v1.0 is an enhanced whole exome capture Panel. It extends the target regions to SNP skeleton of whole genome, intronic regions related to common gene fusion in solid tumors and classical microsatellite loci on the basis of NEXome Core Panel, which spans a 43.3 Mb region of human genome.
- Product details
- Kit Content
- FAQ
- Ordering Information
- Resources
- SNP skeleton of whole genome
- Common gene fusion in solid tumors
Contains intron regions and non-coding regions related to common fusions and can be used to analyze fusion loci in solid tumors.
|
ALK intron 18 - 19 |
BCL2 3'UTR |
BCR intron 8, 13 - 14 |
BRAF intron 7 - 10 |
BRCA1 intron 2, 7 - 8, 12, 16, 19 - 20 |
BRCA2 intron 2 |
CD74 intron 6 - 8 |
EGFR intron 7, 15, 24 - 27 |
ETV4 intron 5 - 6 |
ETV5 intron 6 - 7 |
ETV6 intron 5 - 6 |
|
EWSR1 intron 6 - 13 |
EZR intron 9 - 12 |
FGFR1 intron 1, 5, 17 |
FGFR2 intron 1, 17 |
FGFR3 intron 17 |
FLI1 intron 3 - 8 |
KIT intron 16 |
KMT2A intron 6 - 11 |
MET intron 1, 14 |
MSH2 intron 5 |
MYB intron 14 |
|
MYC intron 1 |
NOTCH2 intron 26 |
NTRK1 intron 8 - 10 |
NTRK2 intron 12, 15 |
NTRK3 intron 13 - 14 |
NUTM1 intron 1 |
PDGFB intron 1 |
PDGFRA intron 7, 9, 11 |
RAF1 intron 4 - 9 |
RARA intron 2 |
RET intron 7 - 11 |
|
ROS1 intron 31 - 35 |
RSPO2 Upstream, 5'UTR, exon 1 - 2, intron 1 |
SDC4 intron 2 |
SLC34A2 intron 4 |
TMPRSS2 intron 1 - 3 |
|
|
|
|
|
|
- Classical microsatellite loci
|
BAT-25 |
BAT-26 |
BAT-40 |
BAT-RII |
NR-21 |
NR-22 |
NR-24 |
NR-27 |
MONO-27 |
D2S123 |
D5S346 |
D17S261 |
D17S520 |
D17S250 |
D18S34 |
|
Performance
Capture Performance
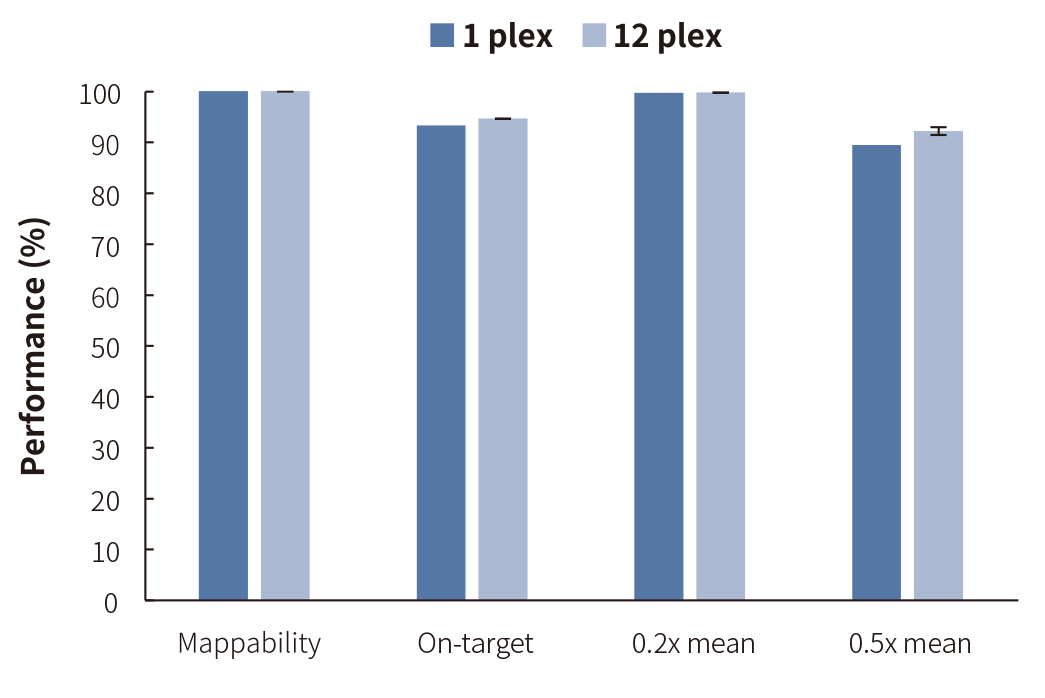
Figure 1. Capture performance of NEXome Plus Panel v1.0 at 1-plex and 12-plex levels. Libraries were prepared using the NadPrep DNA Universal Library Preparation Kit (for Illumina®). NEXome Plus Panel v1.0 were used to complete hybridization capture. Sequencing platform: Illumina HiSeq X Ten, PE150. The BWA was used for alignment to the reference genome hg38, and on-target rate was calculated according to reads number.
Note:The samples: Human Male Genomic DNA (Promega-Male, G1471) and Human Female Genomic DNA (Promega-Female, G1521).
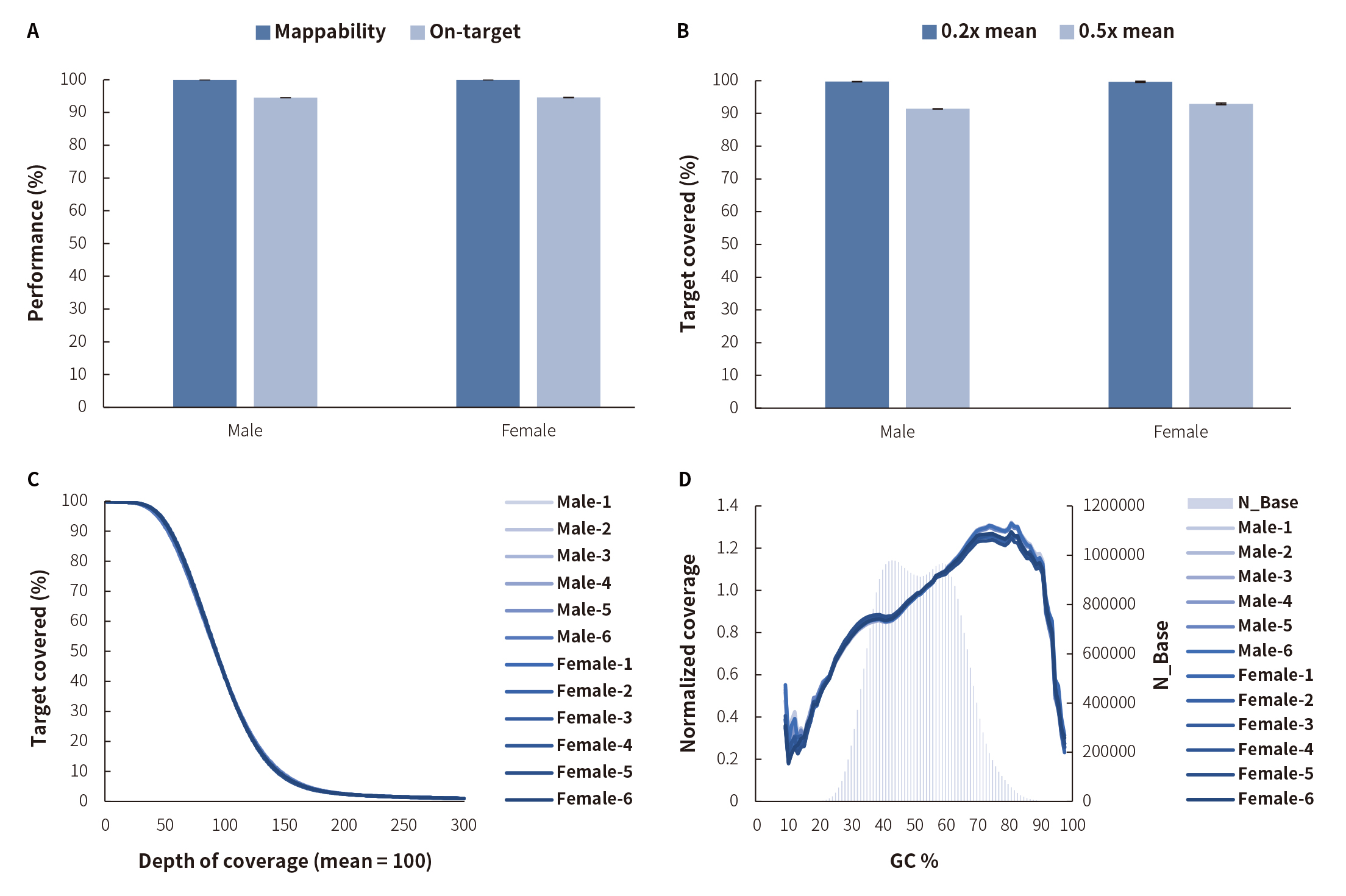
Figure 2. Capture performance of NEXome Plus Panel v1.0 in different gDNA libraries.Libraries were prepared using the NadPrep DNA Universal Library Construction Kit (for Illumina®). NEXome Plus Panel v1.0 (12 plex) were used to complete hybridization capture. Ssequencing platform: Illumina HiSeq X Ten with PE150. The BWA was used for alignment to the reference genome hg38, and on-target rate was calculated according to reads number. A. Mappability and on-target rate; B. Target covered; C. Coverage uniformity and consistency; D. GC bias.
|
Catalog# |
Color of Tube Cap |
Product |
Volume |
Package/Storage |
|
1001861 |
|
NEXome Plus Panel v1.0, 96 rxn |
415 μL |
–20℃ |
|
1001862 |
|
NEXome Plus Panel v1.0, 16 rxn |
70 μL |
–20℃ |
| Product | Catalog# |
| NEXome Plus Panel v1.0, 16 rxn | 1001862 |
| NEXome Plus Panel v1.0, 96 rxn | 1001861 |
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)

Nanodigmbio will contact you within 1 day.
If there are any problems, please contact us by 400 8717 699 / support@njnad.com

