MGI vs Illumina, which μCaler suits you better?
Targeted sequencing based on liquid-phase hybridization capture helps achieve higher sequencing depth at lower costs targeting specific regions of the genome, thereby enhancing detection specificity and sensitivity. However, traditional hybridization capture experiments are not only time-consuming and complex to operation, but they are also significantly influenced by operator handling. Additionally, the capture efficiency for smaller regions is relatively poor. Therefore, traditional hybrid capture is somewhat limited in its application in scenarios that emphasize timeliness and cost sensitivity, such as minimal residual disease (MRD) detection.
To overcome these limitations, Nanodigmbio has released the internationally patented exclusive technology: μCaler Hybrid Capture System, which utilizes probe designs based on the principle of conjugation effect, resulting in more stable probe binding and higher specificity. Additionally, this method also simplifies the operational process, allowing for the completion of the entire hybridization capture process within 2.5 hours. μCaler-series Panel, combined with μCaler Hybrid Capture Reagents v2 and μCaler-series NanoBlockers, constitutes a complete liquid-phase hybridization capture system. It supports comprehensive hybridization capture solutions for both MGI and Illumina mainstream sequencing platforms.
Nanodigmbio's internationally pioneering μCaler Hybrid Capture System, featuring exclusive patented μCaler technology, aims to deliver users with a new experience of precise, stable, efficient, rapid, and straightforward hybridization capture sequencing.
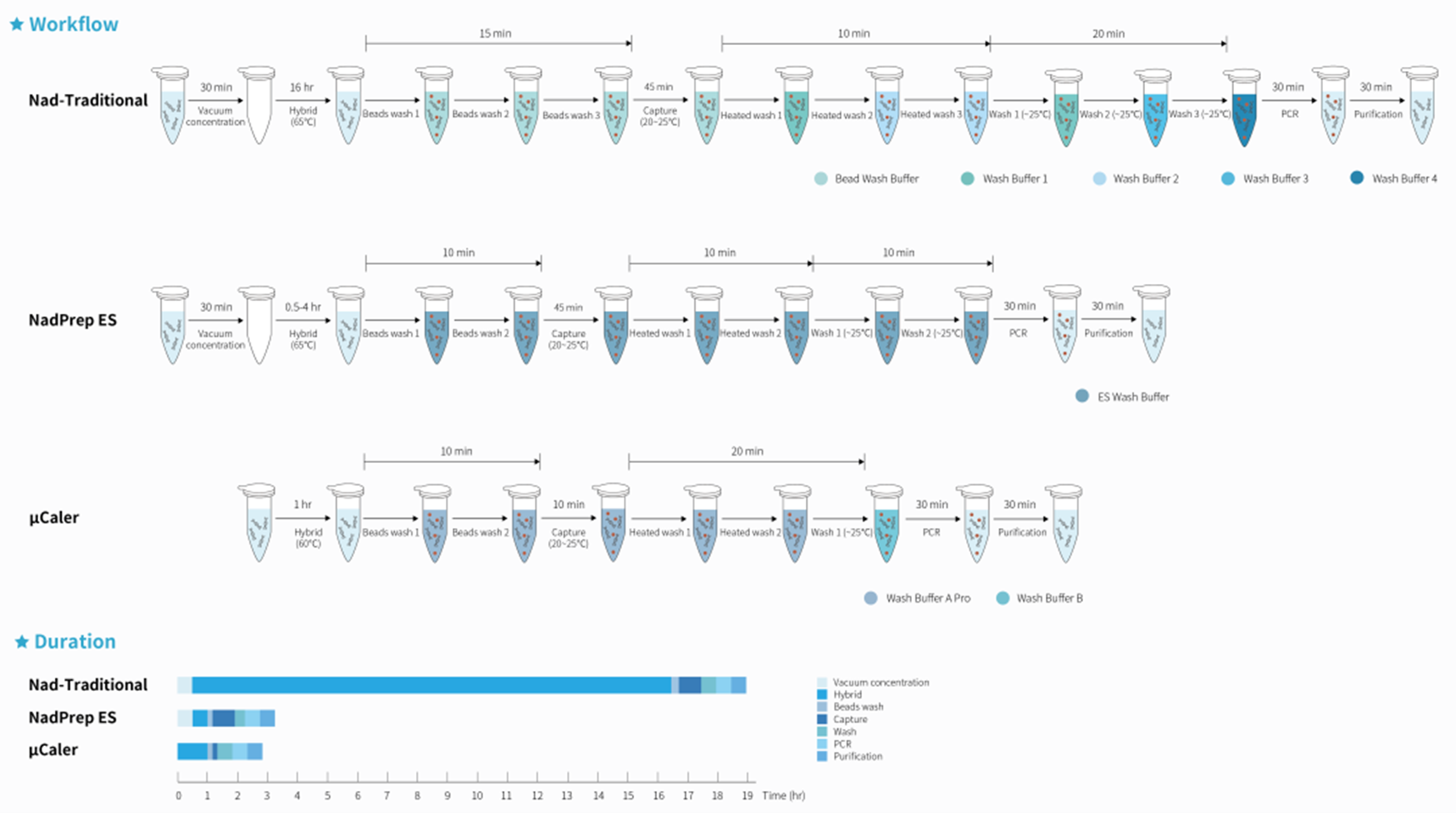
Figure 1. Complete workflow of μCaler Hybrid Capture System.
2.1 μCaler NanoBlockers for IlluminaμCaler NanoBlockers (for Illumina®) are optimized blockers for Illumina® platforms based on μCaler Hybrid Capture System. The μCaler NanoBlockers (for Illumina®) act by reducing non-specific binding of adapter sequences, which improves on-target rate and enrichment efficiency. μCaler NanoBlockers (for Illumina®) work with Illumina libraries with single/dual indexes and 6 nt/8 nt/10 nt indexes.
2.2 μCaler NanoBlockers for MGI
μCaler NanoBlockers (for MGI, DI) are optimized blockers for MGI platforms based on μCaler Hybrid Capture System. The μCaler NanoBlockers (for MGI, DI) facilitates better binding of the library's adapter with 10 nt dual indexes (DI) sequences to the MGI sequencing platform. This reduces non-specific binding between adapters, resulting in improved on-target rates and increased data utilization. 2.3 Comprehensive Solution of μCaler Hybrid Capture System
Nanodigmbio has launched validated solutions for mainstream sequencing platforms: MGI and Illumina, including MRD detection of solid tumor and acute myeloid leukemia (AML), , pathogen microorganism detection, common cancer hotspot mutation detection, and DNA methylation detection.
Table 1. Comprehensive Solution of μCaler Hybrid Capture System.

03 Performance
3.1 Consistent and Reliable Capture across MGI and Illumina PlatformsTo assess the consistency of capture performance between the MGI and Illumina platforms, we conducted multiple comparative tests across both sequencing platforms. The results demonstrate consistent performance in parameters such as mappability, on-target rate, and coverage uniformity (Figure 2).
 Figure 2. Capture performance of μCaler on MGI and Illumina platforms. The pre-libraries were prepared from fragmented human genomic DNA (Promega-male, G1471) by using NadPrep DNA Library Preparation Kit/Module (for Illumina®️/for MGI). 500 ng of each pre-library were pooled for hybridization capture by using μCaler NanoBlockers (for Illumina®) and μCaler NanoBlockers (for MGI, DI) coupled with a 7.5 Kb Panel respectively. Using BWA to alignment to the reference genome (hg19) and On-target rate was calculated by the number of reads. A. Mappability and On-target rate; B. Target covered; C. Fold 80 base penalty.
Figure 2. Capture performance of μCaler on MGI and Illumina platforms. The pre-libraries were prepared from fragmented human genomic DNA (Promega-male, G1471) by using NadPrep DNA Library Preparation Kit/Module (for Illumina®️/for MGI). 500 ng of each pre-library were pooled for hybridization capture by using μCaler NanoBlockers (for Illumina®) and μCaler NanoBlockers (for MGI, DI) coupled with a 7.5 Kb Panel respectively. Using BWA to alignment to the reference genome (hg19) and On-target rate was calculated by the number of reads. A. Mappability and On-target rate; B. Target covered; C. Fold 80 base penalty.Note: Sequencing mode is Illumina Novaseq 6000, PE 150 and DNBSEQ-T7, PE 150 respectively.
3.2 Excellent and Stable Capture Performance
The stable performance of the product not only provides users with the best user experience but also reduces data wastage and lowers sequencing costs. Using reagents with high stability can maximize labor savings and efficiently complete testing tasks accurately. Conversely, instability may result in retesting, leading to increased time and labor costs.
When performing hybrid capture with panels of different sizes and types, the mappability can all reach above 99%, and the on-target rate remain above 60%. In terms of target covered, the 0.2x mean depth of coverage can exceed 99%, and there is high stability observed among different replicates of the same panel.

Figure 3. Stable capture performance of μCaler panels with different sizes. The pre-libraries were prepared from fragmented human genomic DNA (Promega-male, G1471) by using NadPrep DNA Library Preparation Module (for MGI) with NadPrep Universal Adapter (MDI) Module (for MGI). 500 ng of each pre-library were pooled for hybridization capture with three panels of different sizes, ranging from 5 Kb to 42.5 Kb, according to the user manual of μCaler Hybrid Capture of DNA libraries. A. Mappability and On-target rate; B. Target covered.
Note: The sequencing mode is DNBSEQ-T7, PE 150.
3.3 Supports Flexible Hybridization Times
Compared to traditional liquid-phase hybrid capture technologies, the probes in μCaler technology not only bind to target regions but also exhibit a "hand in hand" conjugation effect between probes, allowing for a significant reduction in hybridization time. The test results on the Illumina platform demonstrated that the μCaler Hybrid Capture System exhibits stable performance across different hybridization times. To verify its performance on the MGI platform, we conducted tests with different hybridization times of 1 hr, 2 hr, and 16 hr. The results showed that the capture performance of the μCaler Hybrid Capture System on the MGI platform was consistent across different hybridization times, with optimal performance achieved with a hybridization time of 1 hr or more. This indicates that the μCaler Hybrid Capture System offers flexibility in hybridization time, enabling reliable hybridization effects in a relatively short time, thus improving experimental efficiency while ensuring data stability and quality.

Figure 4. μCaler supports different hybridization time. The pre-libraries were prepared from fragmented human genomic DNA (Promega-male, G1471) by using NadPrep cfDNA Library Preparation Kit (for MGI). Hybridization capture was performed according to the user manual of μCaler Hybrid Capture of DNA libraries using 7.5 Kb and 25 Kb panels, respectively. A. Capture performance across different hybridization times with the 7.5 Kb panel; B. Capture performance across different hybridization times with the 25 Kb panel.
Note: The sequencing mode is DNBSEQ-T7, PE 150.
3.4 Supports Hybridization with Multiple Libraries
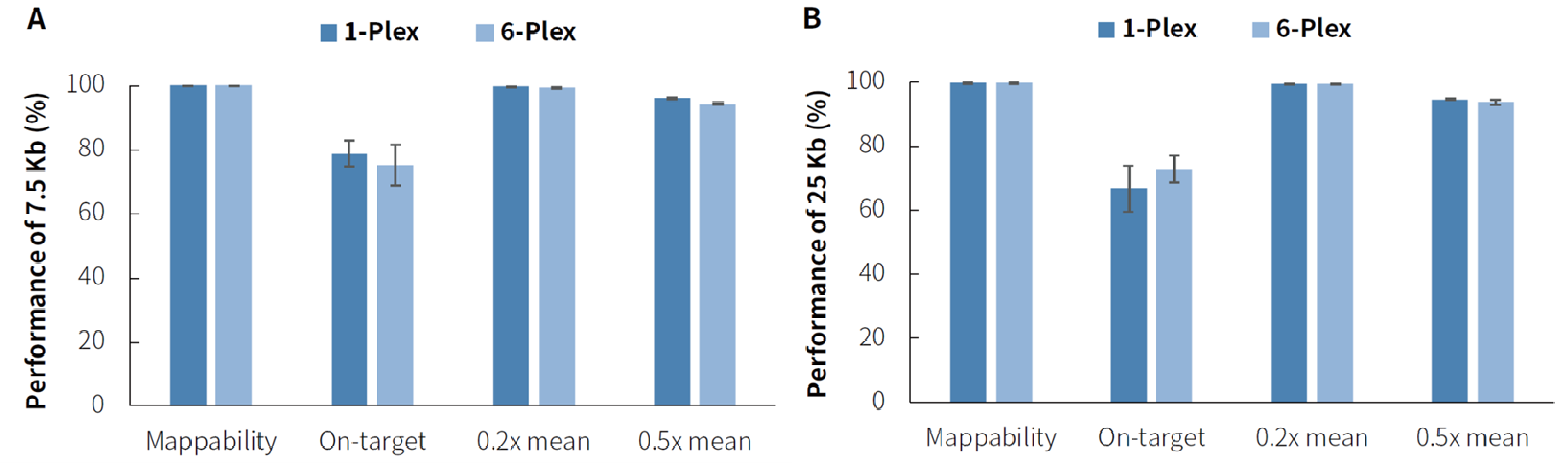
In liquid-phase hybridization capture, the performance of mixed hybridization with multiple libraries is an important criterion for evaluating the performance of reagents. Mixed hybridization not only reduces the cost of reagents but also shortens the experimental duration, saving time and costs. Two different-sized panels (7.5 Kb and 25 Kb) were tested with 500 ng per library (1-plex) and 3,000 ng per 6 libraries (6-plex, 500 ng per library). The results showed that, 6-plex was no significant difference in performance compared to 1-plex, and it still exhibited stable performance. This indicates that the μCaler Hybrid Capture System supports library input amounts ranging from 500 ng to 3000 ng when applied to the MGI platform.
3.5 AML MRD Detection
One of the primary applications of the μCaler Hybrid Capture System is in Minimal Residual Disease (MRD) detection. For the Illumina platforms, we have launched comprehensive MRD solutions of solid tumor and AML. We conducted tests using human genomic DNA (Promega-male, G1471), hematologic disorder gDNA control (LDT Bioscience, LDT-831B), and clinical samples following the μCaler AML MRD Comprehensive Solution. As shown in Figure 6., in terms of basic quality control, the three different samples exhibited consistently excellent performance in mappability, on-target rate, target covered, and so forth. For the hematologic disorder gDNA control, all mutation sites were consistently detected. For clinical samples, the observed allele frequencies were consistent between MGI and Illumina platforms.
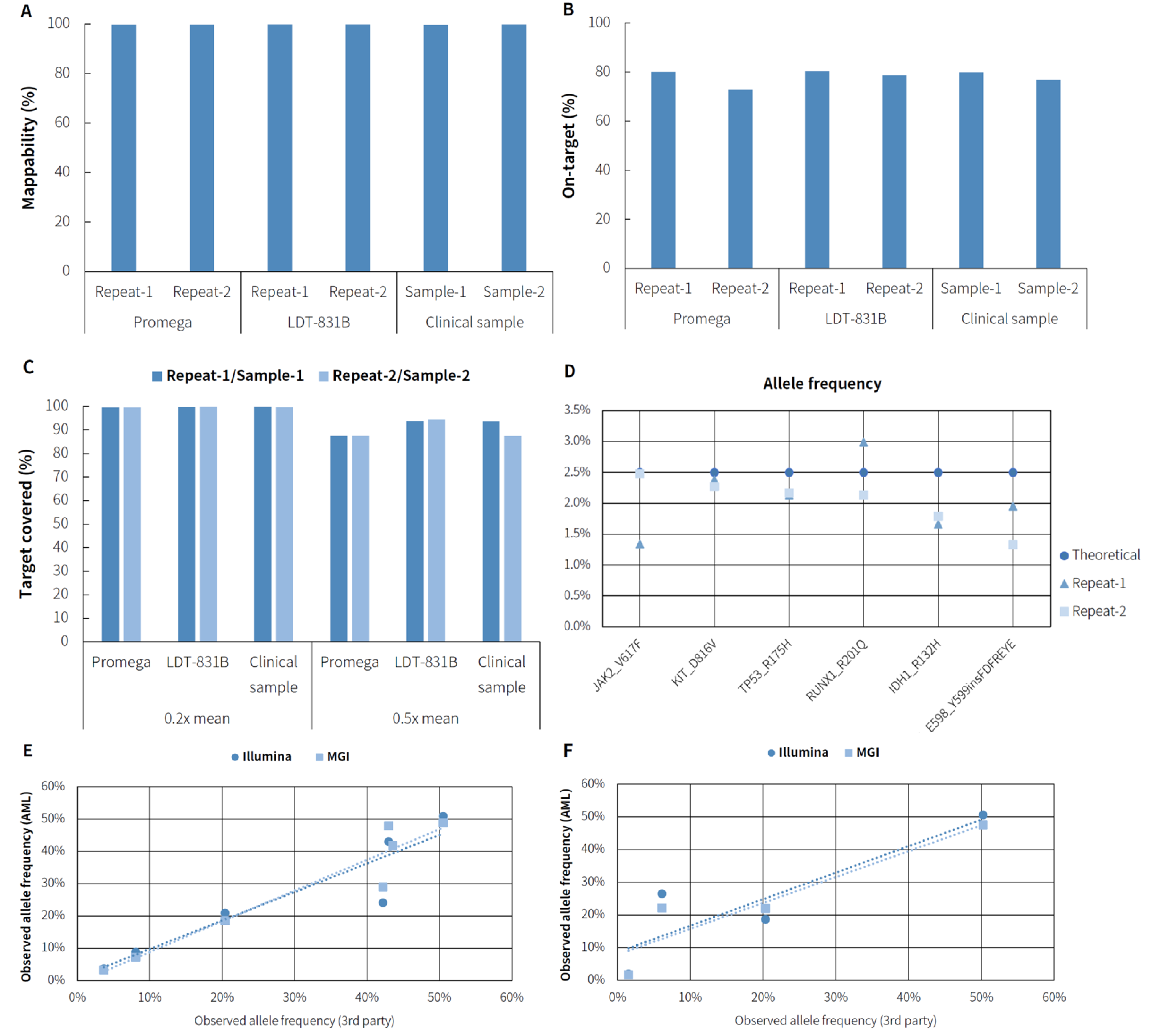
Note: Sequencing mode is Illumina Novaseq 6000, PE 150 and DNBSEQ-T7, PE 150 respectively.
3.6 Hotspot Mutation Detection
For the "hotspot" regions of tumor mutation, Nanodigmbio previously launched μCaler Hotspot Panel v1.0, which involves 49 mutation hotspots in oncogenes and and tumor suppressor genes. This panel demonstrates excellent performance on the Illumina platform. Here, we validated its performance on the MGI platform using the OncoOne Pan-Cancer gDNA Standard (LDT Bioscience, LDT900). As shown in Figure 7., the capture data showed stable mappability, on-target rate, and target covered. No mutation was detected in the negative control, while the observed mutation sites and allele frequencies in the positive control were consistent with the theoretical values of the standard.
 Figure 7. Performance of μCaler HotSpot Panel v1.0 on the MGI platform. Pre-library was prepared using NadPrep EZ DNA Library Preparation Module v2, followed by hybrid capture with μCaler HotSpot Panel v1.0, and variant analysis was conducted using Vardict 1.5.1. A. Basic capture quality control parameters; B. Consistency analysis of standard variation frequency.
Figure 7. Performance of μCaler HotSpot Panel v1.0 on the MGI platform. Pre-library was prepared using NadPrep EZ DNA Library Preparation Module v2, followed by hybrid capture with μCaler HotSpot Panel v1.0, and variant analysis was conducted using Vardict 1.5.1. A. Basic capture quality control parameters; B. Consistency analysis of standard variation frequency.
As an internationally patented exclusive technology, the μCaler Hybrid Capture System demonstrates stable and excellent performance when capturing mini panels on both the MGI and Illumina sequencing platforms. It supports flexible hybridization times and library hybridization modes. Currently, Nanodigmbio has developed various comprehensive solutions in multiple applications based on the μCaler technology, including MRD detection, pathogenic microorganism detection, DNA methylation detection, etc., aiming to contribute to the field of companion diagnostics.
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)


