DNA + RNA Dual-Workflow Synergy for Comprehensive and Precise Detection of Hematologic Malignancy
Hematologic malignancies exhibit high heterogeneity, with different subtypes showing significant variations in their molecular genetic features. These variations include base substitutions, insertions/deletions, copy number variations (CNVs), and gene fusions. Consequently, comprehensive and precise genetic testing is crucial for the diagnosis, treatment strategy formulation, and prognostic evaluation of hematologic malignancies.
At the DNA level, detection covering both coding regions and portions of intronic regions enables the comprehensive identification of point mutations, indels, CNVs, and fusion breakpoints, making it suitable for screening known and potential pathogenic variants. In contrast, RNA-level detection focuses directly on fusion genes at the transcript level, enabling the precise identification of actual fusion events and their partner genes, as well as the assessment of whether these fusions result in aberrant transcripts that impact gene expression. By integrating both DNA and RNA workflows, a dual safeguard is achieved that enhances overall detection comprehensiveness and result accuracy.
To comprehensively decode the mutation landscape of hematologic malignancies and enhance fusion gene detection, Nanodigmbio has launched NanoHema Panel v2.0 —— a dual-detection panel that integrates both DNA and RNA analysis for hematologic malignancies.
01 Introduction
NanoHema Panel v2.0 targets 481 genes associated with hematologic malignancies, comprehensively covering a wide range of genetic alterations including point mutations, insertions/deletions, gene amplifications, and gene fusions.
This panel consists of two components:
NanoHema Panel v2.0_DNA Tube: Covers 1.7 Mb of genomic regions, including full coding regions of 436 genes, partial intronic regions of 12 genes, and the IGH Switch regions.
NanoHema Panel v2.0_RNA Tube: Covers 146 genes, optimized for fusion gene detection.
By integrating multi-omics approaches, employing a highly targeted design, and enhancing clinical utility, NanoHema Panel v2.0 offers an efficient and cost-effective solution for precision diagnosis, treatment planning, and longitudinal management of hematologic malignancies.

Figure 1. Gene list covered by NanoHema Panel v2.0.
02 Feature
Comprehensive Multi-type Mutation Coverage
• Encompasses 481 hematologic malignancy-related genes, enabling thorough analysis of base substitutions, indels, amplifications, and gene fusions.
Dual Workflow Synergy for Enhanced Accuracy
• RNA workflow compensates for the limitations of the DNA workflow by accurately identifying fusion gene partners and validating the functional impact of fusion events. The synergistic use of both workflows ensures comprehensive and precise detection.
Flexible Customization
• Supports flexible subdivision into multiple sub-panels or the addition of target genes, allowing adaptation to diverse hematologic malignancy diagnostic needs in both research and clinical applications.
03 Performance
3.1 NanoHema Panel v2.0_DNA
3.1.1 Stable Capture Performance
NanoHema Panel v2.0_DNA was used for hybrid capture on various types of gDNA samples and sequenced on multiple platforms. The results demonstrated outstanding performance across key metrics: Mappability exceeding 99.9%, an average On-target rate of over 91% (Figure 2.A), with 0.2x mean and 0.5x mean coverage reaching above 99% and 94% respectively (Figure 2.B); the Fold 80 value was maintained below 1.5 (Figure 2.C), and the average sequencing depth approached 500x (Figure 2.D).
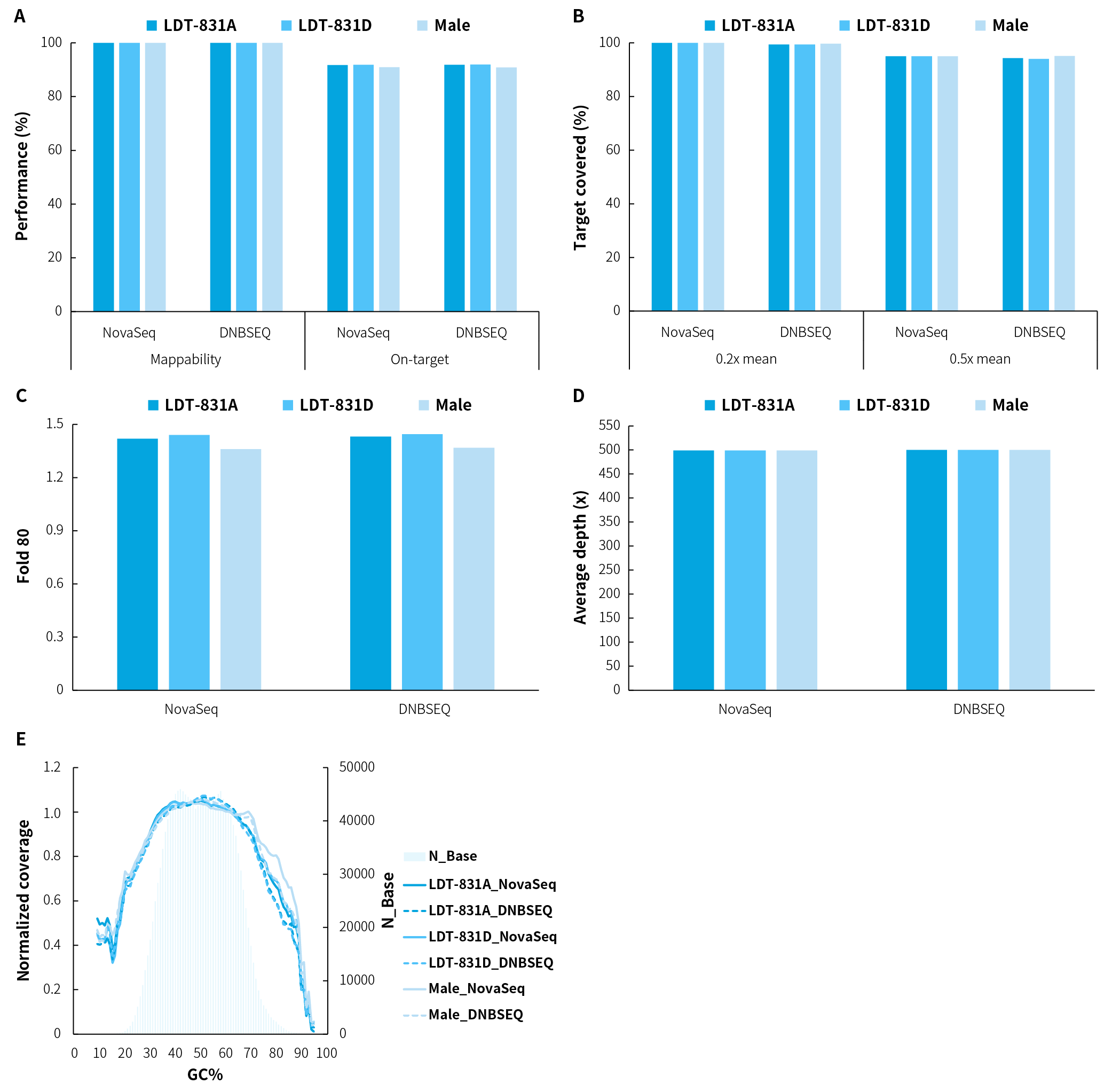
Figure 2. Capture performance of NanoHema Panel v2.0_DNA applied to various types of gDNA samples. A. Mappability & On-target rate; B. Target covered; C. Fold 80 base penalty; D. Average sequencing depth after deduplication; E. GC bias. Using 50 ng of various gDNA samples, pre-libraries were prepared with the NadPrep EZ DNA Library Preparation Kit coupled with the NadPrep Universal Stubby Adapter (UDI) Module, followed by hybrid capture using NanoHema Panel v2.0_DNA with NadPrep Hybrid Capture Reagents. Sequencing was performed on both Novaseq 6000 (PE150) and DNBSEQ-T7 (PE150) platforms, with a random subset of 1.3 Gb used for data analysis.
Note: LDT-831: hematologic malignancy gDNA quality control (LDT Bioscience; LDT-831A: AF = 5%; LDT-831D: AF = 0%). Male: human genomic DNA standard (Promega, G1471).
3.1.2 Sensitive Detection of Multiple Mutation Types
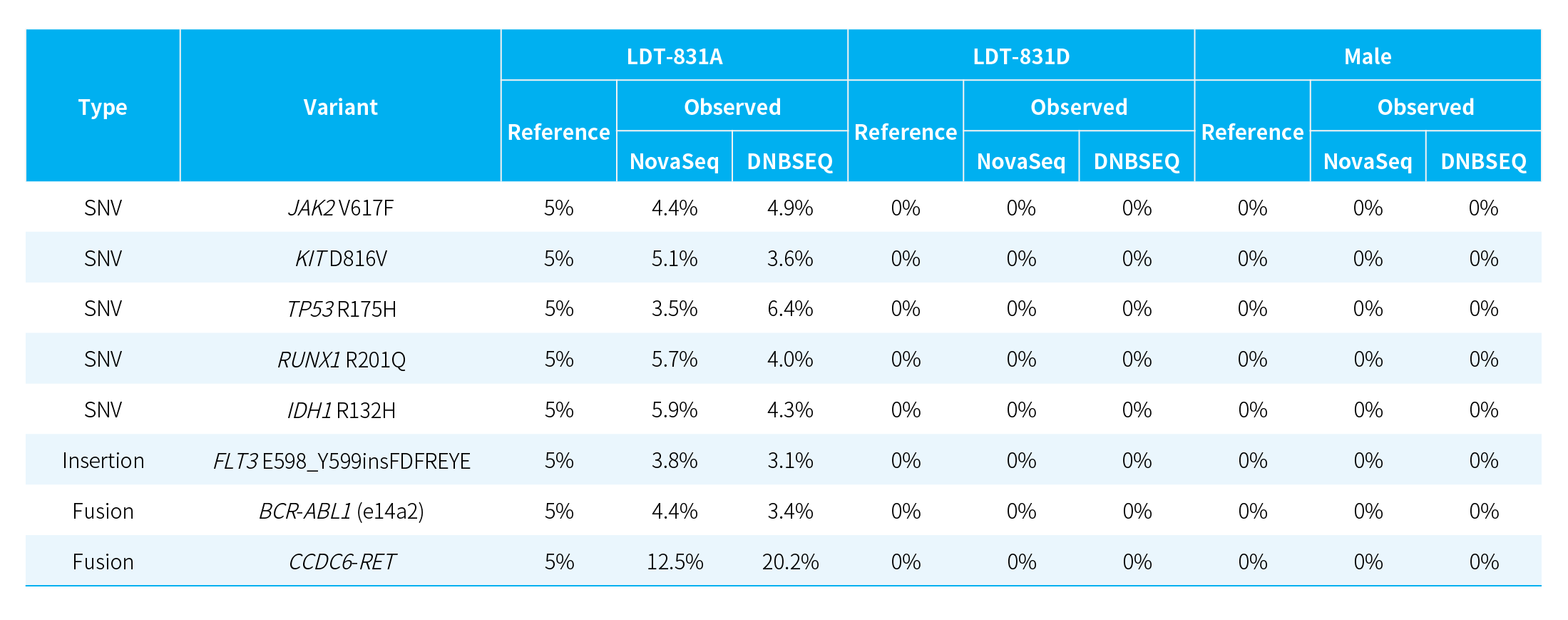
Using gDNA quality controls with various mutation levels, NanoHema Panel v2.0_DNA was evaluated for its mutation detection performance. The results showed a high concordance between the observed variant allele frequencies and the reference values provided by the quality controls.
Table 1. Detection of various mutation types in different standards using NanoHema Panel v2.0_DNA.
3.2 NanoHema Panel v2.0_RNA
3.2.1 Efficient On-target Capture
Pre-libraries prepared from K562 cell line RNA were
hybrid captured to assess the on-target performance of NanoHema Panel v2.0_RNA.
The results demonstrated that the panel achieved an on-target rate exceeding
96% across different sequencing platforms (Figure 3).
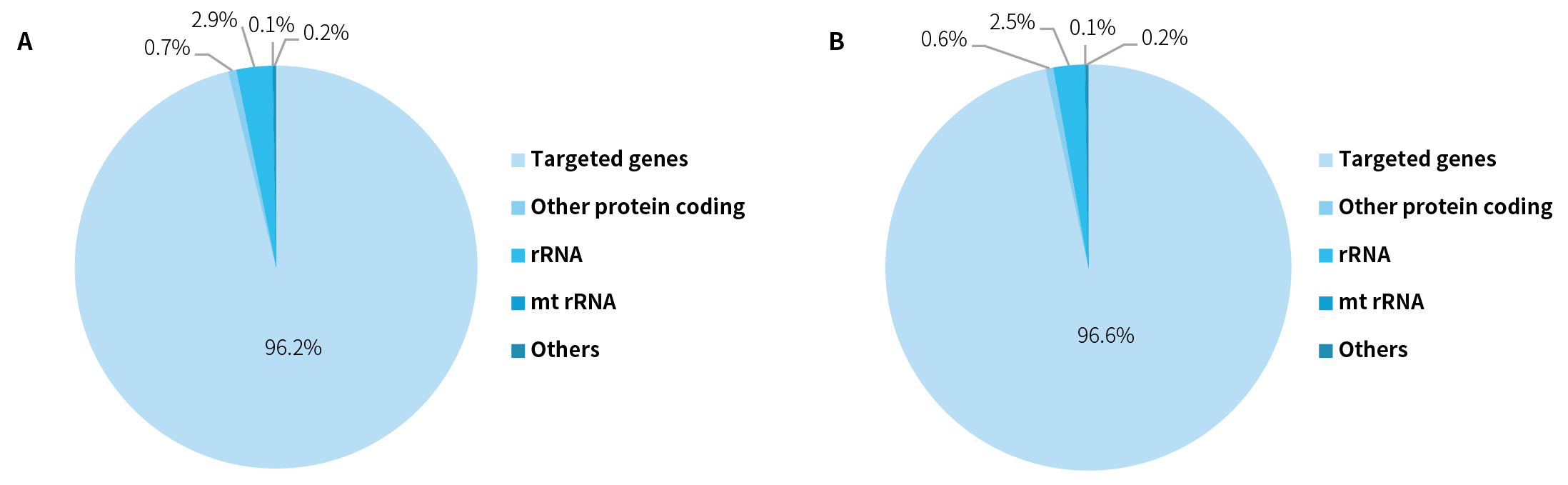
Figure 3. Composition of capture sequencing data using NanoHema Panel v2.0_RNA. For the K562 cell line RNA (50 ng), pre-libraries were prepared using the NadPrep Total RNA-To-DNA Module coupled with the NadPrep DNA Library Preparation Kit v2, followed by hybrid capture with NanoHema Panel v2.0_RNA and NadPrep Hybrid Capture Reagents. Sequencing was performed on Novaseq 6000 (PE150) (A) and DNBSEQ-T7 (PE150) (B), with analysis performed using Salmon.
3.2.2 Precise Fusion Detection
As a highly efficient multiplex amplification library preparation kit, this product enables enrichment of the CDR3 region in the human immune repertoire within as little as 3 hours. Users can flexibly select IG- or TR-specific enrichment modes based on research objectives, thereby reducing sequencing data volume while enhancing sensitivity for low-frequency clone detection. Whether applied to basic immune repertoire profiling, clonality analysis in lymphoid malignancies, or minimal residual disease (MRD) monitoring, this kit provides robust technical support for both research and clinical applications, facilitating efficient execution of complex immunological studies!
Table 2. Detection of fusion genes in K562 cell line RNA using NanoHema Panel v2.0_RNA.
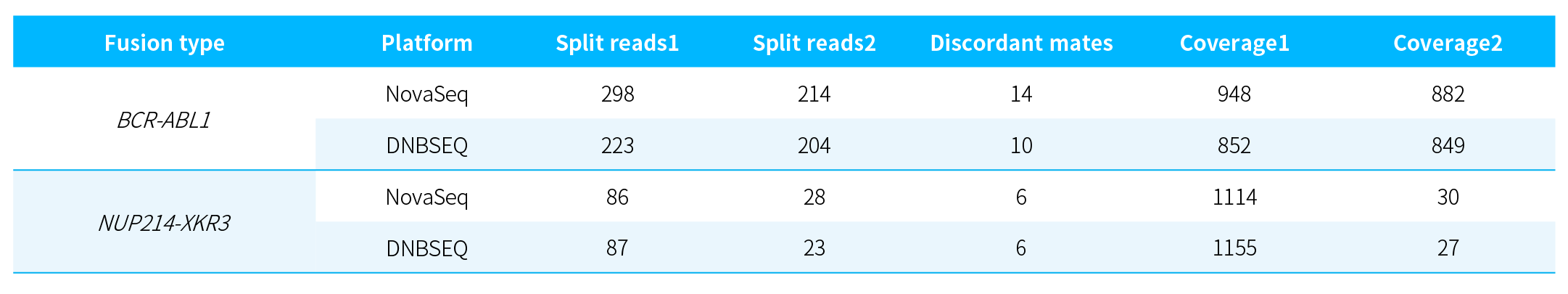
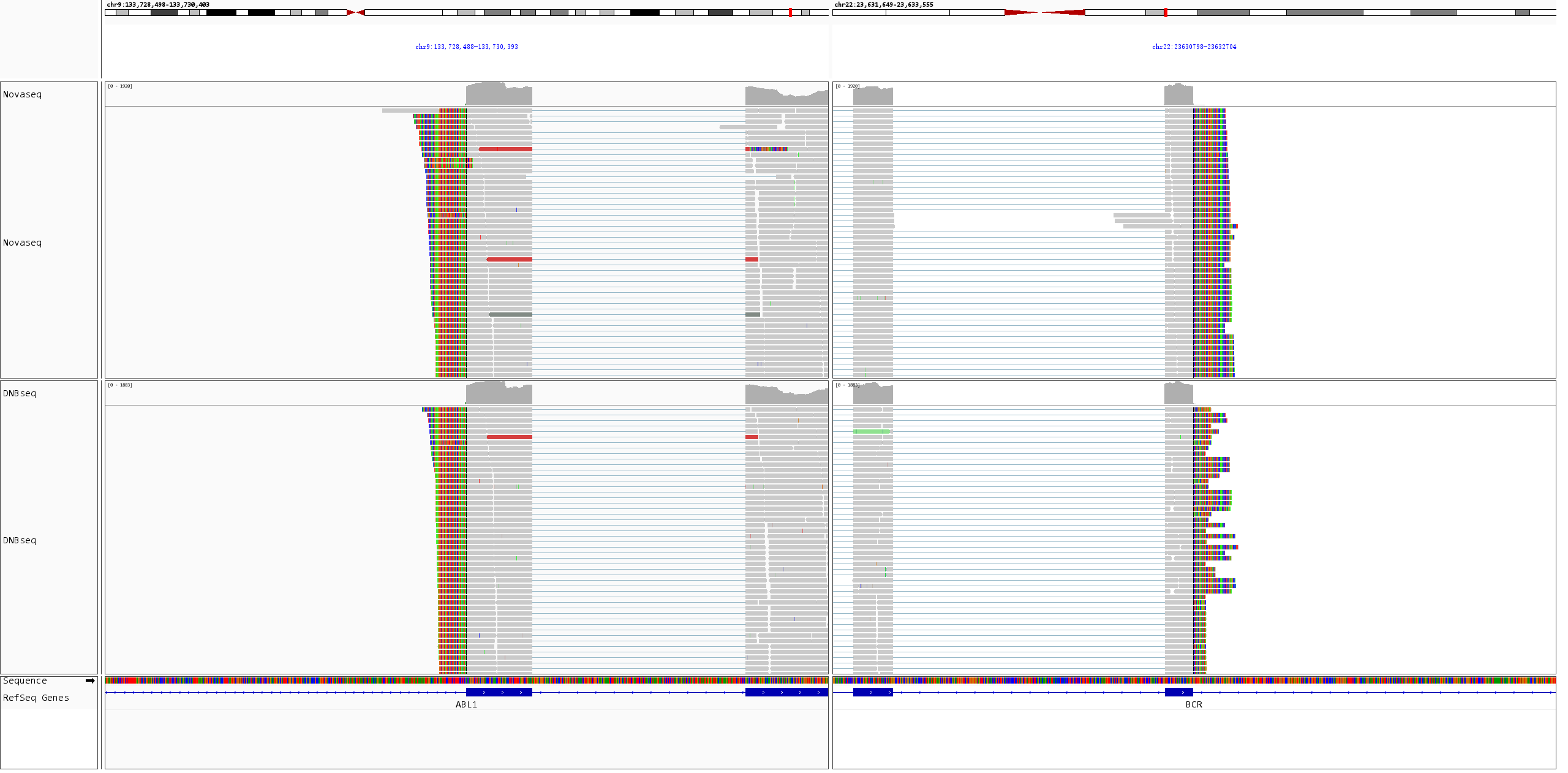
Fusion events in K562 cell line RNA were visualized using IGV. As shown in Figure 4, both BCR-ABL1 and NUP214-XKR3 fusion events were successfully detected, further validating the reliability of the results.
A
B
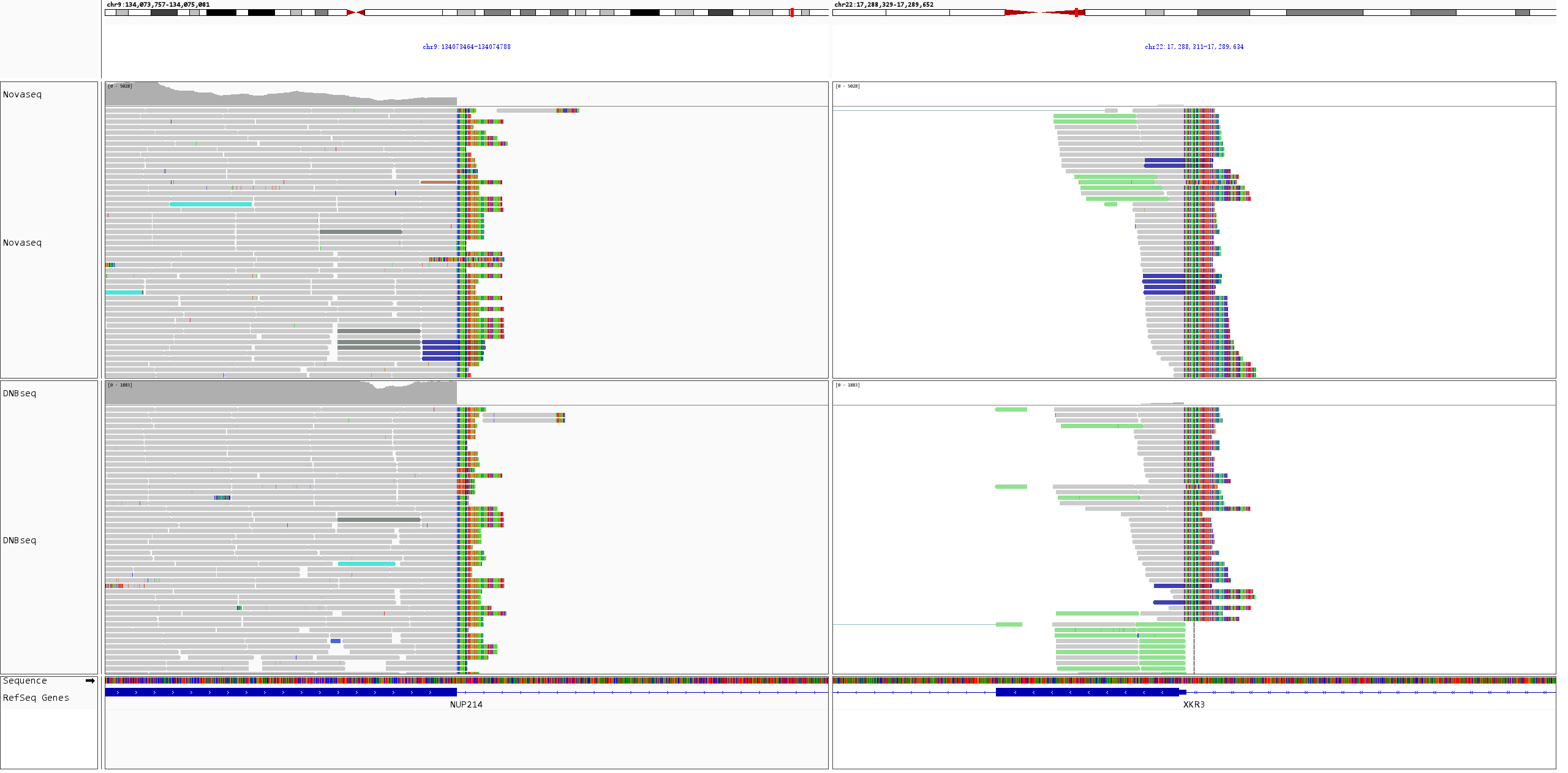
Figure 4. IGV visualization of fusion events in K562 cell line RNA. A. BCR-ABL1 fusion; B. NUP214-XKR3 fusion.
04 Applications and Prospects
NanoHema Panel v2.0 integrates a synergistic DNA and RNA dual workflow, enabling multidimensional analysis of genetic alterations. DNA detection provides broad coverage for mutation screening, while RNA detection precisely confirms fusion expression and functionality, collectively ensuring comprehensive and accurate results. This multi-omics integration strategy not only enhances clinical reliability through cross-validation but also reveals hidden rearrangements that are often missed by conventional single-omics approaches.
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)