
DMD Research Panel v1.0
#1001892
DMD Research Panel v1.0 targets the DMD gene
that encodes dystrophin, covering an approximately 2.2 Mb genomic region for
enrichment and comprehensive analysis of the entire DMD gene sequence.
- Product details
- Kit Content
- FAQ
- Ordering Information
- Resources
Performance
Basic Quality Control Performance on Dual Platforms
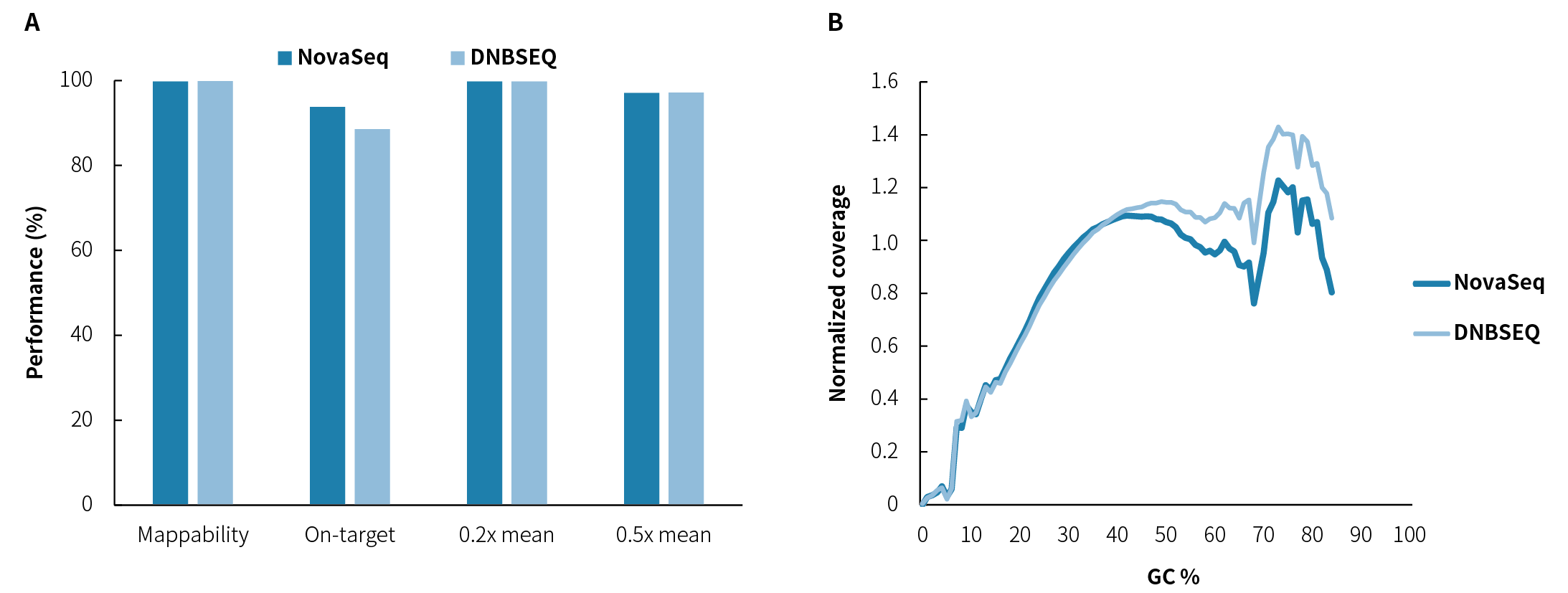
Figure 1. Basic quality control performance of the DMD Research Panel v1.0. A. Mappability, On-target rate, and Target covered; B. GC bias. Pre-library preparation was performed using the NadPrep EZ DNA Library Preparation Kit with the NadPrep Universal Stubby Adapter (UDI) Module, followed by hybrid capture using the DMD Research Panel v1.0 and NadPrep Hybrid Capture Reagents. Sequencing was performed on NovaSeq 6000 (PE150) and DNBSEQ-T7 (PE150).
Note: Samples were human genomic DNA standard (Promega, G1471).
Capture Performance on DMD gDNA Reference Standards
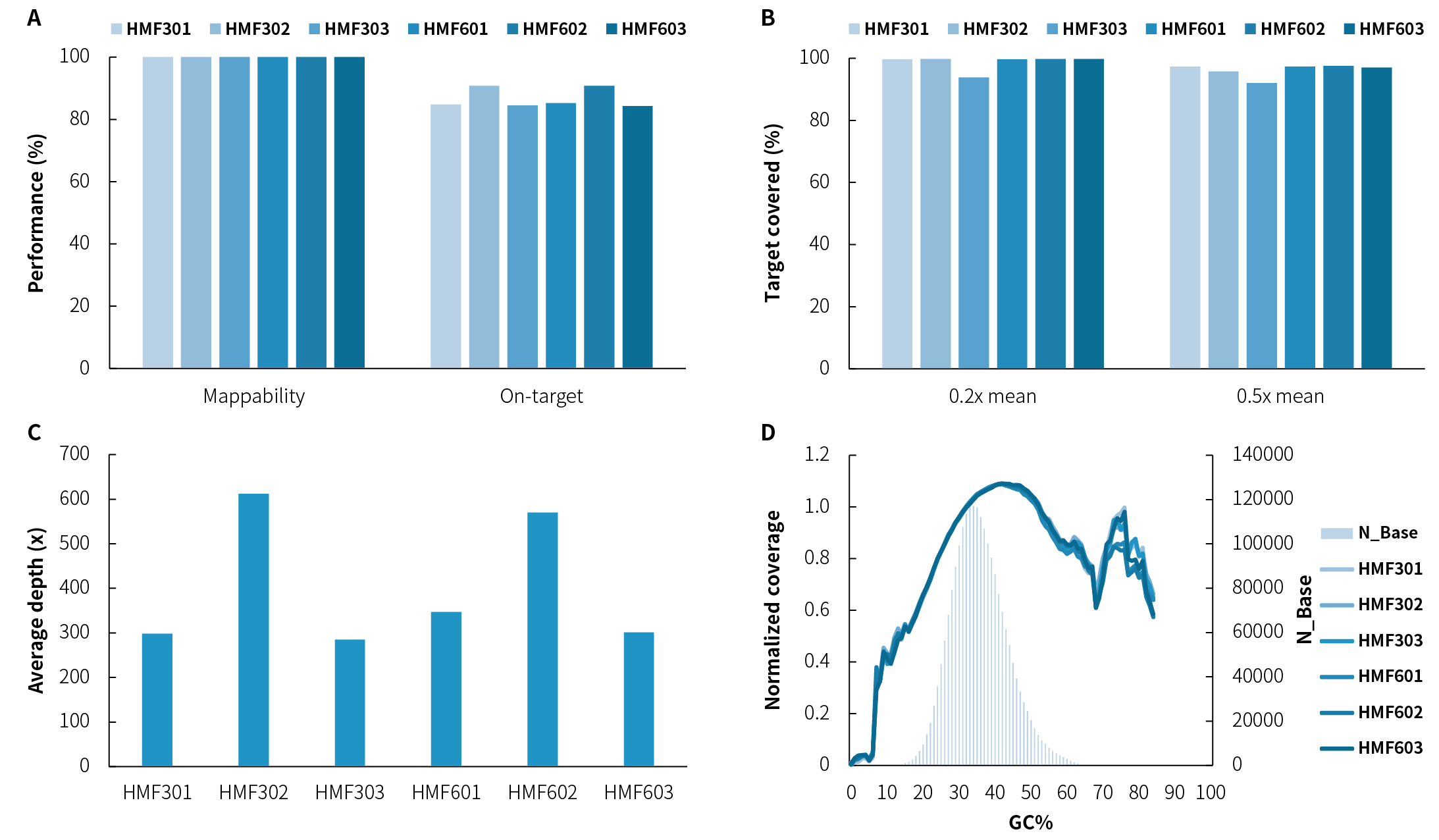
Figure 2. Capture performance of the DMD Research Panel v1.0 on reference standards. A. Mappability & On-target rate; B. Target covered; C. Average sequencing depth (without deduplication); D. GC bias. Sequencing was performed on NovaSeq 6000 (PE150). Data of 0.78, 1.5, 0.75, 0.9, 1.4, and 0.79 Gb were used for analysis.
Note: Samples are derived from DMD gDNA Reference Standards (GeneWell). HMF301-3 correspond to GW-HMF301-3, and HMF601-3 correspond to GW-HMF601-3; within the two families, the gender of standards 1-3 are Male, Female, and Male, respectively; Genotypes of HMF301-3 by MLPA are Normal, Exon48-Exon50 heterozygous deletion and Exon48-Exon50 deletion/hemizygote, while genotypes for HMF601-3 are Normal, Exon18-Exon25 haplox repeat, and Exon18-Exon25 repeat.
Precise Detection of CNV Breakpoints
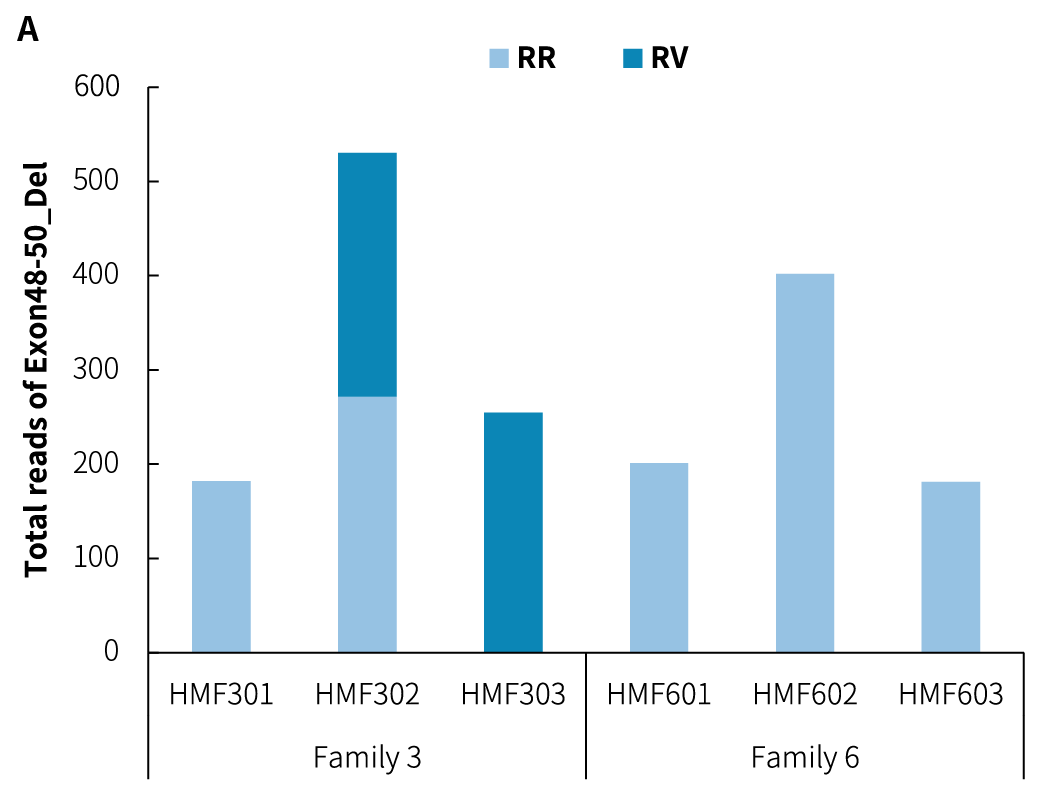
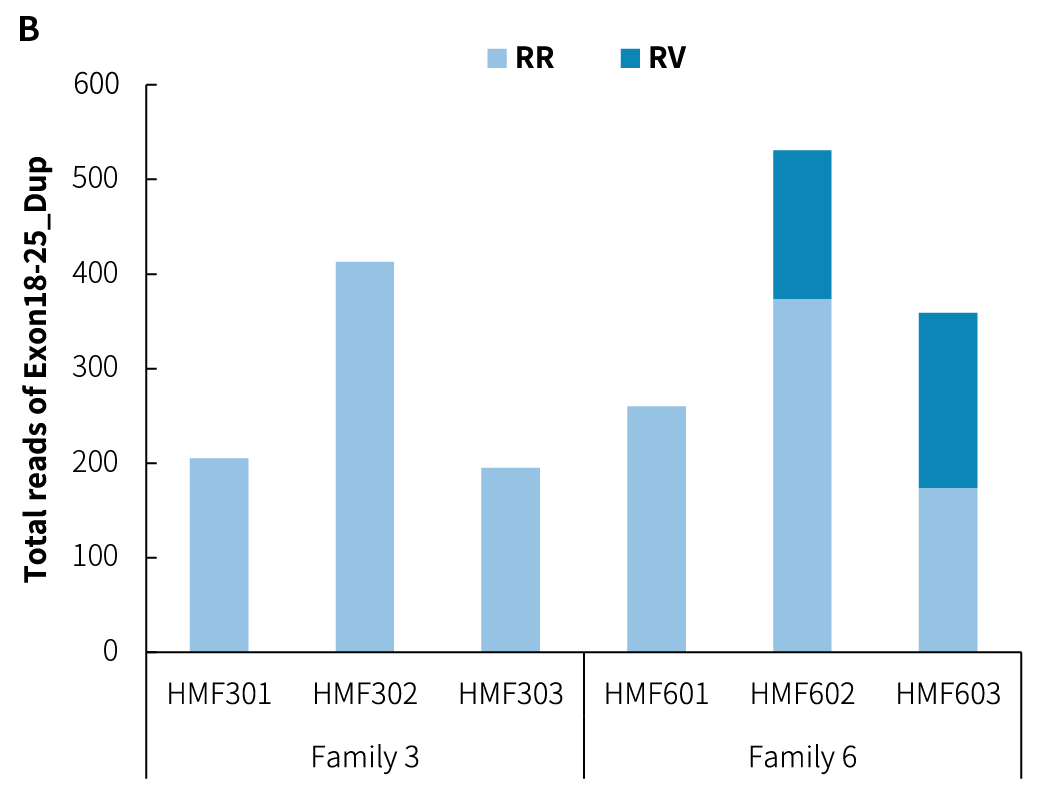
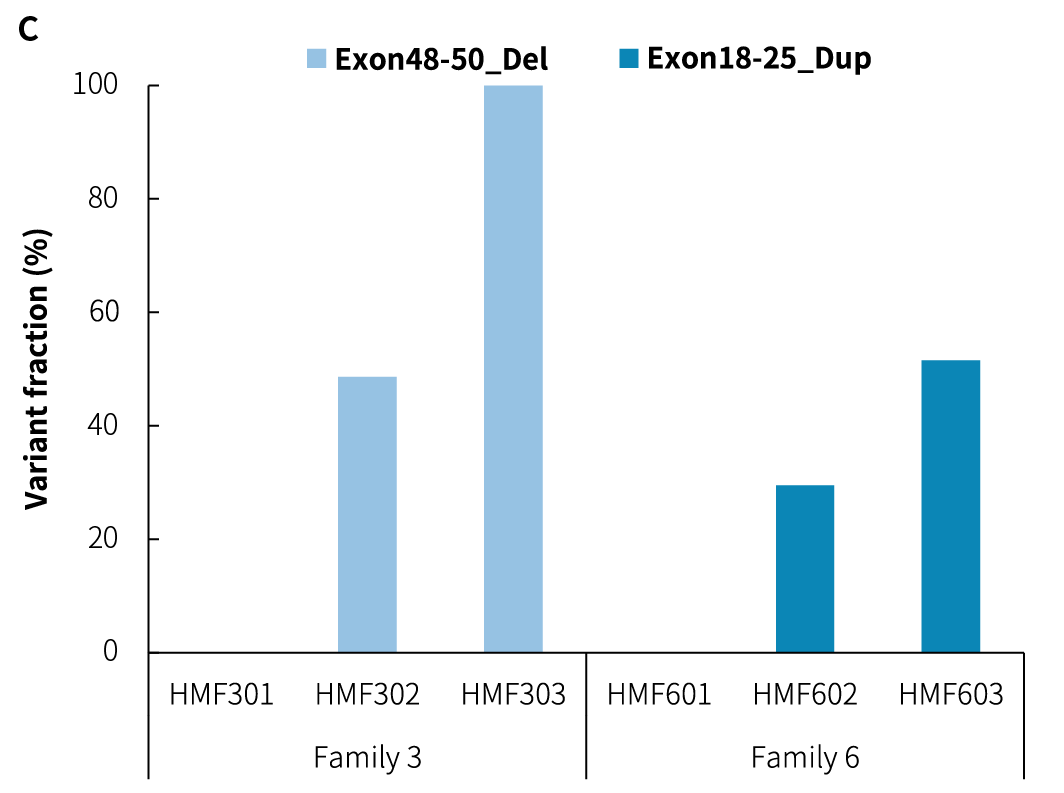
Figure 3. Detection of CNV breakpoints in reference standards using the DMD Research Panel v1.0. A. Analysis of Exon48-50 deletion (Del) reads; B. Analysis of Exon18-25 duplication (Dup) reads; C. Variant fraction. Sequencing data were aligned to the hg38 reference genome using BWA, and variant analysis was performed using Delly to count the supporting reads.
Note: RR: reference junction reads; RV: variant junction reads; Exon48-50_Del: deletion of Exon48-Exon50; Exon18-25_Dup: duplication of Exon18-Exon25.
DMD Genomic Coverage
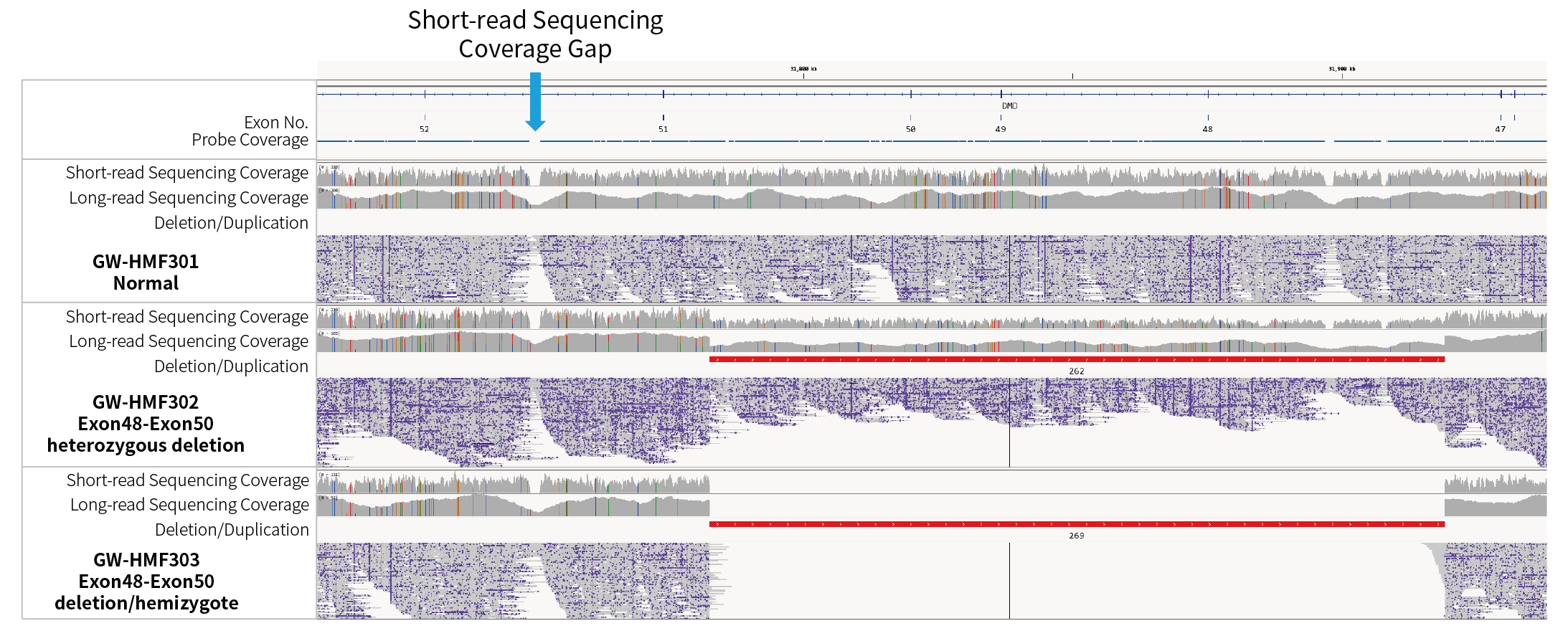
Figure 4. Genomic coverage of DMD gDNA Reference Standards after targeted capture with the DMD Research Panel v1.0, as assessed by short-read (NovaSeq 6000, PE150) and long-read (Oxford Nanopore) sequencing.
| Catalog# | Cap Color | Item | Volume | Package/Storage |
| 1001891 |
|
DMD Research Panel v1.0, 96 rxn | 415 μL | -25~-15℃ |
| 1001892 |
|
DMD Research Panel v1.0, 16 rxn | 70 μL | -25~-15℃ |
| Product | Catalog# |
| DMD Research Panel v1.0, 96 rxn | 1001891 |
| DMD Research Panel v1.0, 16 rxn | 1001892 |
Product Sheet
Application Note
Demo Data
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)

Nanodigmbio will contact you within 1 day.
If there are any problems, please contact us by 400 8717 699 / support@njnad.com



