No Need for Conversion: Ultra-sensitive Solution for Synchronous Detection of Methylation and Mutation in a Single Reaction Coming Soon!
View: 1796 / Time: 2024-08-01
DNA methylation is a chemical covalent modification. Without changing the DNA sequence, it affects the activity of DNA fragments and thereby alters the genetic manifestation. It is a highly conserved epigenetic modification and is widely present in the genomes of eukaryotes such as plants and animals. It plays a crucial role in biological processes, affecting regulated gene expression, embryonic development, cell proliferation and differentiation. At the same time, it serves a significant function in maintaining genomic stability and resisting the invasion of exogenous DNA viruses.
Abnormal DNA methylation in cancer cells is involved throughout the entire process of cancer initiation and progression. Compared with traditional tumor markers, DNA methylation has the advantages of being earlier, less invasive, and more accurate. Therefore, detecting methylation markers can assist in the early diagnosis of cancer, assess the risk of progression, assess minimal residual disease (MRD) of tumors and monitor recurrence. It can also serve as a marker of chemotherapy sensitivity to better guide the treatment plan[1-5].
01 Methylation Conversion: "Stumbling Block" for Multi-omics Analysis
The detection of DNA methylation usually requires conversion processing, with mainstream technologies including bisulfite (BS) conversion based on chemical reagents and mild conversion based on enzymatic (EM) methods.
With the treatment of BS or EM, the unmethylated cytosine (C) can be converted into uracil (U). After PCR amplification, it further becomes thymine (T), while the originally methylated C remains unchanged. This allows for the differentiation of methylation states after sequencing. Although BS conversion is fast, efficient, and has a short processing time, the drastic temperature and pH changes during conversion can cause DNA degradation and fragmentation. Therefore, despite being regarded as the "gold standard" for DNA methylation detection, BS conversion is more suitable for samples with a higher initial amount.
The relatively mild EM conversion methods reduced DNA degradation and fragmentation, but the operation is cumbersome, the reaction time is long, and the conversion efficiency is not as stable as the BS conversion method, requiring further optimization.
DNA methylation detection based on NGS offers high throughput, making it suitable for the detection of single cancer type, multiple cancer types, or pan-cancer types. However, limited by the low initial amount of cfDNA samples and the need for simultaneous mutation detection, the operation of multi-omics analysis is complex and the high costs (Figure 1.). The common library conversion strategies on the market, such as "library preparation before conversion" or "conversion before library preparation", are difficult to achieve the desired effect.
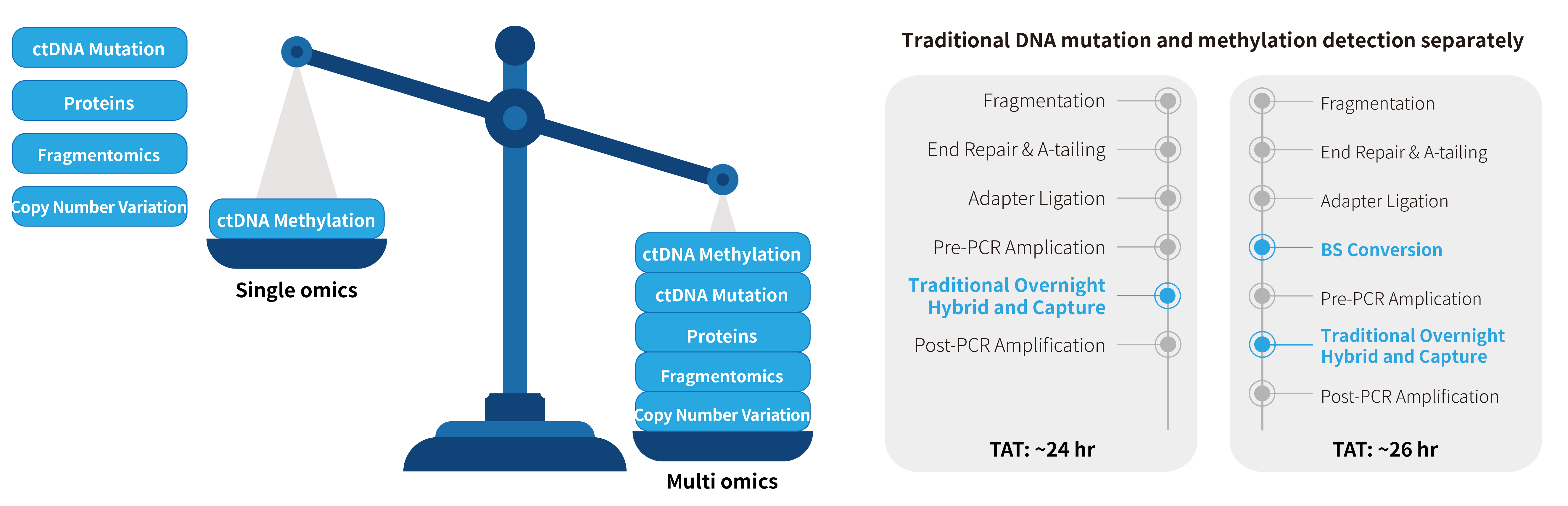
Figure 1. Process of the traditional enrichment technical scheme for separate detection of methylation and mutation.
How to achieve the synchronous detection of methylation and mutation signals from a single and limited sample in a manner that is both easy to operate and retains the original sequence information while achieving higher sensitivity is a pressing challenge in multi-omics analysis of the current liquid biopsy technology.
02 Synchronous Detection of methylation and mutation without Sequence Conversion
Based on the methylation sensitive restriction endonuclease (MSRE) technology, which utilizes the principle that MSRE cannot cleave sequences containing methylated modifications in the site, there is no need for sequence conversion. Therefore, this method requires a low amount of initial sample and offers high alignment efficiency, showing significant potential for development[6].
Combining the MSRE technology and the exclusive patented ultra-sensitive probe hybridization capture system (μCaler™ Hybrid System), Nanodigmbio is about to officially launch the ultra-sensitive, comprehensive solution for the synchronous detection of methylation and mutation without the need for sequence conversion (μCaler™ DNA Full Screen System). This solution only requires one initial sample, one hybridization reaction, and one set of capture probes to achieve an ultra-highly sensitive panoramic detection of methylation and mutation!
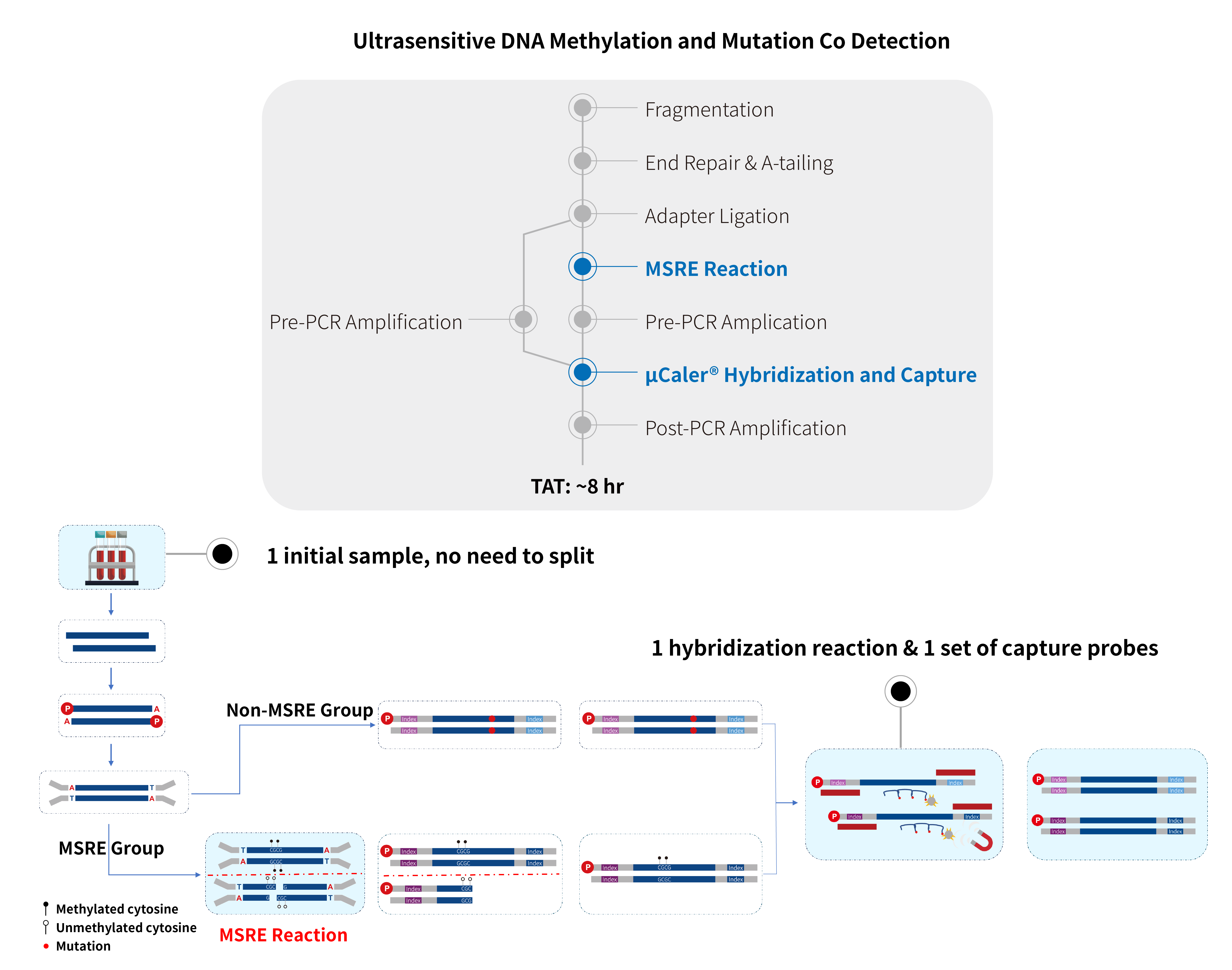
Figure 2. μCaler™ DNA Full Screen System efficiently achieves synchronous of methylation and mutation signals.
03 Features of μCaler DNA Full Screen System
• Enhanced Data Utilization: Abandoning the traditional BS conversion solution, fully retains the richness of the original sequence information, thereby enhancing data utilization;
• Single-process Multi-omics Analysis: Requires only one sample, one hybridization reaction, and one set of capture probes to perform multi-omics analysis;
• High-speed and Convenience: Simplified and rapid workflow allows for the efficient simultaneous detection of methylation and mutation signals within a single day;
• Combined Analysis to Capture Weak Signals: Increases the detection sensitivity of low-abundance methylation and ultra-low-frequency mutation signals;
• High Stability: Higher capture efficiency and stability, offering more uniform coverage;
• Powerful Innovation: A probe design solution with independent intellectual property rights and core competitiveness;
• End-to-End Comprehensive Solution: Covers the entire process from library preparation, hybridization capture, to bioinformatics data analysis, providing professional probe design and personalized bioinformatics analysis solutions.
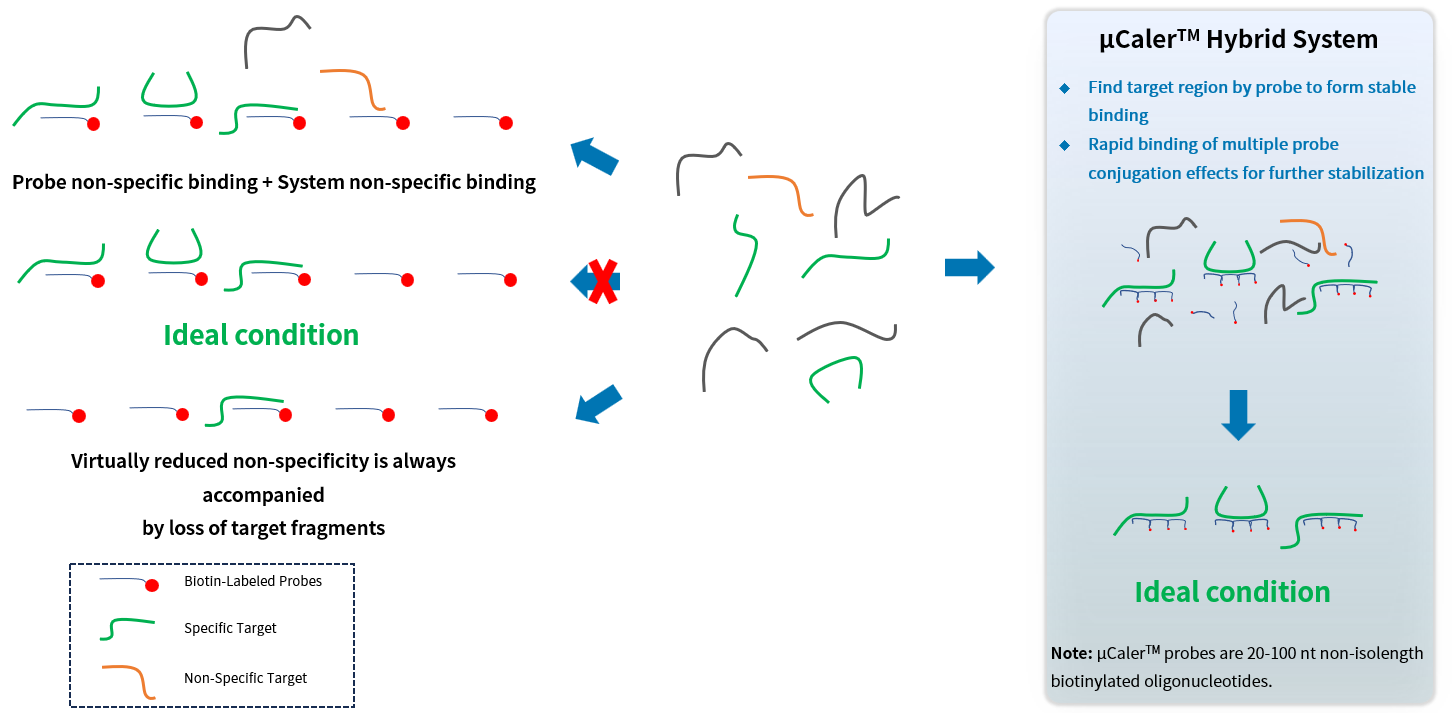
Ultra-sensitive Probe Hybridization Capture System (μCaler™ Hybrid System)
Compared with the traditional liquid-phase hybridization capture technology, Nanodigmbio's globally exclusive patented μCaler™ Hybrid System uses non-isolength probes (20 - 100 nt). Through the conjugation effect between the probes, it enhances the enrichment capability of adjacent probes for the target region, efficiently and stably capturing the methylation and mutation signals in the sample to achieve multi-omics analysis.
References
[1] Papanicolau-Sengos A, Aldape K. DNA methylation profiling: an emerging paradigm for cancer diagnosis[J]. Annual Review of Pathology: Mechanisms of Disease, 2022, 17(1): 295-321.
[2] LIAO Leen, PAN Zhizhong. Comprehensive diagnosis and treatment of precision surgery for colorectal cancer [J]. Chinese Journal of Oncology Prevention and Treatment, 2023, 15(4): 361-369.
[3] Jin S, Zhu D, Shao F, et al. Efficient detection and post-surgical monitoring of colon cancer with a multi-marker DNA methylation liquid biopsy[J]. Proceedings of the National Academy of Sciences, 2021, 118(5): e2017421118.
[4] Shen S Y, Singhania R, Fehringer G, et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes[J]. Nature, 2018, 563(7732): 579-583.
[5] Vessies D C L, Schuurbiers M M F, van der Noort V, et al. Combining variant detection and fragment length analysis improves detection of minimal residual disease in postsurgery circulating tumour DNA of stage II–IIIA NSCLC patients[J]. Molecular oncology, 2022, 16(14): 2719-2732.
[6] Tumor marker committee of Chinese anti-cancer association. Expert Consensus on detection and clinical application of tumor DNA methylation markers (Version 2024)[J]. Chinese Journal of Oncology Prevention and Treatment, 2024, 16(2): 129-142.
Abnormal DNA methylation in cancer cells is involved throughout the entire process of cancer initiation and progression. Compared with traditional tumor markers, DNA methylation has the advantages of being earlier, less invasive, and more accurate. Therefore, detecting methylation markers can assist in the early diagnosis of cancer, assess the risk of progression, assess minimal residual disease (MRD) of tumors and monitor recurrence. It can also serve as a marker of chemotherapy sensitivity to better guide the treatment plan[1-5].
01 Methylation Conversion: "Stumbling Block" for Multi-omics Analysis
The detection of DNA methylation usually requires conversion processing, with mainstream technologies including bisulfite (BS) conversion based on chemical reagents and mild conversion based on enzymatic (EM) methods.
With the treatment of BS or EM, the unmethylated cytosine (C) can be converted into uracil (U). After PCR amplification, it further becomes thymine (T), while the originally methylated C remains unchanged. This allows for the differentiation of methylation states after sequencing. Although BS conversion is fast, efficient, and has a short processing time, the drastic temperature and pH changes during conversion can cause DNA degradation and fragmentation. Therefore, despite being regarded as the "gold standard" for DNA methylation detection, BS conversion is more suitable for samples with a higher initial amount.
The relatively mild EM conversion methods reduced DNA degradation and fragmentation, but the operation is cumbersome, the reaction time is long, and the conversion efficiency is not as stable as the BS conversion method, requiring further optimization.
DNA methylation detection based on NGS offers high throughput, making it suitable for the detection of single cancer type, multiple cancer types, or pan-cancer types. However, limited by the low initial amount of cfDNA samples and the need for simultaneous mutation detection, the operation of multi-omics analysis is complex and the high costs (Figure 1.). The common library conversion strategies on the market, such as "library preparation before conversion" or "conversion before library preparation", are difficult to achieve the desired effect.

Figure 1. Process of the traditional enrichment technical scheme for separate detection of methylation and mutation.
How to achieve the synchronous detection of methylation and mutation signals from a single and limited sample in a manner that is both easy to operate and retains the original sequence information while achieving higher sensitivity is a pressing challenge in multi-omics analysis of the current liquid biopsy technology.
02 Synchronous Detection of methylation and mutation without Sequence Conversion
Based on the methylation sensitive restriction endonuclease (MSRE) technology, which utilizes the principle that MSRE cannot cleave sequences containing methylated modifications in the site, there is no need for sequence conversion. Therefore, this method requires a low amount of initial sample and offers high alignment efficiency, showing significant potential for development[6].
Combining the MSRE technology and the exclusive patented ultra-sensitive probe hybridization capture system (μCaler™ Hybrid System), Nanodigmbio is about to officially launch the ultra-sensitive, comprehensive solution for the synchronous detection of methylation and mutation without the need for sequence conversion (μCaler™ DNA Full Screen System). This solution only requires one initial sample, one hybridization reaction, and one set of capture probes to achieve an ultra-highly sensitive panoramic detection of methylation and mutation!

Figure 2. μCaler™ DNA Full Screen System efficiently achieves synchronous of methylation and mutation signals.
03 Features of μCaler DNA Full Screen System
• Enhanced Data Utilization: Abandoning the traditional BS conversion solution, fully retains the richness of the original sequence information, thereby enhancing data utilization;
• Single-process Multi-omics Analysis: Requires only one sample, one hybridization reaction, and one set of capture probes to perform multi-omics analysis;
• High-speed and Convenience: Simplified and rapid workflow allows for the efficient simultaneous detection of methylation and mutation signals within a single day;
• Combined Analysis to Capture Weak Signals: Increases the detection sensitivity of low-abundance methylation and ultra-low-frequency mutation signals;
• High Stability: Higher capture efficiency and stability, offering more uniform coverage;
• Powerful Innovation: A probe design solution with independent intellectual property rights and core competitiveness;
• End-to-End Comprehensive Solution: Covers the entire process from library preparation, hybridization capture, to bioinformatics data analysis, providing professional probe design and personalized bioinformatics analysis solutions.
Ultra-sensitive Probe Hybridization Capture System (μCaler™ Hybrid System)
Compared with the traditional liquid-phase hybridization capture technology, Nanodigmbio's globally exclusive patented μCaler™ Hybrid System uses non-isolength probes (20 - 100 nt). Through the conjugation effect between the probes, it enhances the enrichment capability of adjacent probes for the target region, efficiently and stably capturing the methylation and mutation signals in the sample to achieve multi-omics analysis.

In the subsequent chapters, we will present more application cases and the performance of the ultra-sensitive synchronous detection comprehensive solution for methylation and mutation (μCaler™ DNA Full Screen System). Stay tuned!
Welcome to contact sales@njnad.com or local sales for more information.
References
[1] Papanicolau-Sengos A, Aldape K. DNA methylation profiling: an emerging paradigm for cancer diagnosis[J]. Annual Review of Pathology: Mechanisms of Disease, 2022, 17(1): 295-321.
[2] LIAO Leen, PAN Zhizhong. Comprehensive diagnosis and treatment of precision surgery for colorectal cancer [J]. Chinese Journal of Oncology Prevention and Treatment, 2023, 15(4): 361-369.
[3] Jin S, Zhu D, Shao F, et al. Efficient detection and post-surgical monitoring of colon cancer with a multi-marker DNA methylation liquid biopsy[J]. Proceedings of the National Academy of Sciences, 2021, 118(5): e2017421118.
[4] Shen S Y, Singhania R, Fehringer G, et al. Sensitive tumour detection and classification using plasma cell-free DNA methylomes[J]. Nature, 2018, 563(7732): 579-583.
[5] Vessies D C L, Schuurbiers M M F, van der Noort V, et al. Combining variant detection and fragment length analysis improves detection of minimal residual disease in postsurgery circulating tumour DNA of stage II–IIIA NSCLC patients[J]. Molecular oncology, 2022, 16(14): 2719-2732.
[6] Tumor marker committee of Chinese anti-cancer association. Expert Consensus on detection and clinical application of tumor DNA methylation markers (Version 2024)[J]. Chinese Journal of Oncology Prevention and Treatment, 2024, 16(2): 129-142.
Solutions
- Methyl Library Preparation Total Solution
- Sequencing single library on different platform--Universal Stubby Adapter (UDI)
- HRD score Analysis
- Unique Dual Index for MGI platforms
- RNA-Cap Sequencing of Human Respiratory Viruses Including SARS-CoV-2
- Total Solution for RNA-Cap Sequencing
- Total Solution for MGI Platforms
- Whole Exome Sequencing
- Low-frequency Mutation Analysis
Events
-
Exhibition Preview | Nanodigmbio invites you to join us at Boston 2025 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at WHX & WHX Labs Kuala Lumpur 2025, Malaysia International Trade and Exhibition Centre in Kuala Lumpur

-
Exhibition Preview | Nanodigmbio Invites You to Join Us at Hospitalar 2025, Brazil International Medical Device Exhibition in São Paulo

-
Exhibition Preview | Nanodigmbio invites you to join us at Denver 2024 Annual Meeting of the American Society of Human Genetics (ASHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Sapporo 2024 Annual Meeting of the Japan Society of Human Genetics (JSHG)

-
Exhibition Preview | Nanodigmbio invites you to join us at Association for Diagnostics & Laboratory Medicine (ADLM)